---
title: An overview of the R programming environment
author: Deepayan Sarkar
---
```{r opts, echo = FALSE, results = "hide", warning = FALSE, message = FALSE}
opts_chunk$set(dev = "png", cache=TRUE, cache.path='~/knitr-cache/roverview/', autodep=TRUE,
## prompt = TRUE,
comment = "",
warning = TRUE, message = TRUE,
knitr.table.format = "html",
fig.width = 15, fig.height = 8, dpi = 96, fig.path='figures/roverview-')
library(lattice)
lattice.options(default.args = list(as.table = TRUE),
default.theme = list(fontsize = list(text = 20, points = 12)))
options(warnPartialMatchDollar = FALSE, width = 120, show.signif.stars = FALSE)
```
## Software for Statistics
- Computing software is essential for modern statistics
- Large datasets
- Visualization
- Simulation
- Iterative methods
- Many softwares are available
. . .
- We will learn about R
- Available as [Free](https://en.wikipedia.org/wiki/Free_software_movement) /
[Open Source](https://en.wikipedia.org/wiki/The_Open_Source_Definition) Software
- Very popular (both academia and industry)
- Easy to try out on your own
## Outline
- Installing R
- Some examples
- A little bit of history
- Some thoughts on why R has been successful
## Installing R
* R is most commonly used as a [REPL](https://en.wikipedia.org/wiki/Read-eval-print_loop) (Read-Eval-Print-Loop)
* This is essentially the model used by a calculator:
* Waits for user input
* Evaluates and prints result
* Waits for more input
. . .
* There are several different _interfaces_ to do this
* R itself works on many platforms (Windows, Mac, UNIX, Linux)
* Some interfaces are platform-specific, some work on most
. . .
* R and the interface may need to be installed separately
## Installing R
* Go to (or choose a [mirror](https://cran.r-project.org/mirrors.html) first)
* Follow instructions depending on your platform (probably Windows)
. . .
* This will install R, as well as a default graphical interface on Windows and Mac
. . .
* I will recommend a different interface called
[R Studio](https://www.rstudio.com/) that needs to be installed separately
* I personally use yet another interface called
[ESS](https://ess.r-project.org/) which works with a general purpose
editor called [Emacs](https://www.gnu.org/software/emacs/)
([download link](https://vigou3.github.io/emacs-modified-windows/)
for Windows)
## Running R
* Once installed, you can start the appropriate interface (or R
directly) to get something like this:\
\
```
R Under development (unstable) (2018-05-05 r74699) -- "Unsuffered Consequences"
Copyright (C) 2018 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
Natural language support but running in an English locale
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
Loading required package: utils
>
```
* The `>` represents a _prompt_ indicating that R is waiting for input.
* The difficult part is to learn what to do next
## Before we start, an experiment!
![the dress]()
\includegraphics[width=0.4\textwidth]{the_dress.png}
. . .
Color combination: Is it __white & gold__ or __blue & black__ ? Let's count!
```{r echo=FALSE}
NN <- 44
XX <- 26
```
## Question: What proportion of population sees white & gold?
- Statistics uses data to make inferences
- Model:
- Let $p$ be the probability of seeing white & gold
- Assume that individuals are independent
. . .
- Data:
- Suppose $X$ out of $N$ sampled individuals see white & gold;
e.g., $N = `r {NN}`$, $X = `r {XX}`$.
- According to model, $X \sim Bin(N, p)$
. . .
- "Obvious" estimate of $p = X / N = `r {XX}` / `r {NN}` = `r {round(XX/NN, 4)}`$
- But how is this estimate derived?
## Generally useful method: maximum likelihood
- Likelihood function: probability of observed data as function of $p$
$$
L(p) = P(X = `r {XX}`) = {`r {NN}` \choose `r {XX}`} p^{`r {XX}`} (1-p)^{(`r {NN}`-`r {XX}`)}, p \in (0, 1)
$$
- Intuition: $p$ that gives higher $L(p)$ is more "likely" to be correct
- Maximum likelihood estimate $\hat{p} = \arg \max L(p)$
. . .
- By differentiating
$$
\log L(p) = c + `r {XX}` \log p + `r {NN-XX}` \log (1-p)
$$
we get
$$
\frac{d}{dp} \log L(p) = \frac{`r {XX}`}{p} - \frac{`r {NN-XX}`}{1-p} = 0 \implies `r {XX}` (1-p) - `r {NN-XX}` p = 0 \implies p = \frac{`r {XX}`}{`r {NN}`}
$$
## How could we do this numerically?
- Pretend for the moment that we did not know how to do this.
- How could we arrive at the same solution numerically?
- Basic idea: Compute $L(p)$ for various values of $p$ and find minimum.
. . .
- To do this in R, the most important thing to understand is that __R
works like a calculator__:
- The user types in an expression, R calculates the answer
- The expression can involve numbers, variables, and functions
. . .
- For example:
```{r echo=FALSE}
N = NN
x = XX
fakeCode <- sprintf("```r\nN = %d\nx = %d\n```", NN, XX)
```
`r {fakeCode}`
```{r}
p = 0.5
choose(N, x) * p^x * (1-p)^(N-x)
```
## "Vectorized" computations
- One distinguishing feature of R is that it operates on "vectors"
```{r}
pvec = seq(0, 1, by = 0.01)
pvec
Lvec = choose(N, x) * pvec^x * (1-pvec)^(N-x)
Lvec
```
## Plotting is very easy
```{r}
plot(x = pvec, y = Lvec, type = "l")
```
\
## Functions
- Functions can be used to encapsulate repetitive computations
- Like mathematical functions, R function also take arguments as input
and "returns" an output
```{r}
L = function(p) choose(N, x) * p^x * (1-p)^(N-x)
L(0.5)
L(x/N)
```
## Functions can be plotted directly
```{r}
plot(L, from = 0, to = 1)
```
\
## ...and they can be numerically "optimized"
```{r}
optimize(L, interval = c(0, 1), maximum = TRUE)
```
\
- Compare with
```{r}
x / N
```
## A more complicated example
- Suppose $X_1, X_2, ..., X_n \sim Bin(N, p)$, and are independent
- Instead of observing each $X_i$, we only get to know $M = \max(X_1, X_2, ..., X_n)$
- What is the maximum likelihood estimate of $p$? ($N$ and $n$ are known, $M = m$ is observed)
## A more complicated example
To compute likelihood, we need p.m.f. of $M$ :
$$
P(M \leq m) = P(X_1 \leq m, ..., X_n \leq m) =
\left[ \sum_{x=0}^m {N \choose x} p^{x} (1-p)^{(N-x)} \right]^n
$$
and
$$
P(M = m) = P(M \leq m) - P(M \leq m-1)
$$
. . .
In R,
```{r}
n = 10
N = 50
M = 30
F <- function(p, m)
{
x = seq(0, m)
(sum(choose(N, x) * p^x * (1-p)^(N-x)))^n
}
L = function(p)
{
F(p, M) - F(p, M-1)
}
```
## Maximum Likelihood estimate
```{r echo=FALSE,fig.height = 6}
pp = seq(0.2,0.8,by=0.005)
plot(pp, sapply(pp, L), type = "l")
```
\
```{r}
optimize(L, interval = c(0, 1), maximum = TRUE)
```
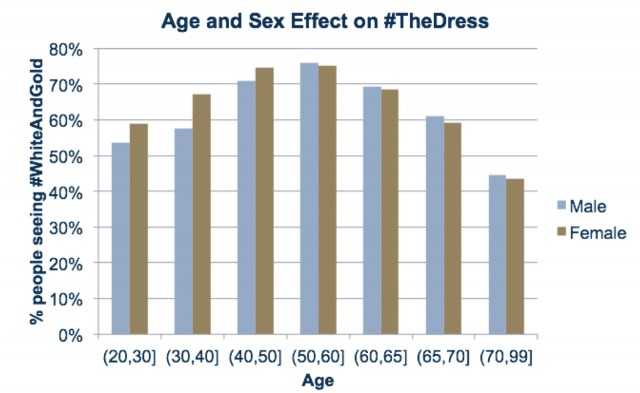
## "The Dress" revisited
- What factors determine perceived color? (From [23andme.com](https://blog.23andme.com/23andme-research/genetics-and-that-striped-dress/))
![age-sex effect]()
\includegraphics[width=0.8\textwidth]{Age-Effect-Dress.jpg}
## Simulation: birthday problem
- R can be used to simulate random events
- Example: how likely is a common birthday in a group of 20 people?
```{r}
N = 20
days = sample(365, N, rep = TRUE)
days
length(unique(days))
```
## Law of Large Numbers
- With enough replications, sample proportion should converge to probability
```{r}
haveCommon = function()
{
days = sample(365, N, rep = TRUE)
length(unique(days)) < N
}
haveCommon()
haveCommon()
haveCommon()
haveCommon()
```
## Law of Large Numbers
- With enough replications, sample proportion should converge to probability
- Do this sytematically:
```{r}
replicate(100, haveCommon())
```
## Law of Large Numbers
- With enough replications, sample proportion should converge to probability
```{r}
plot(cumsum(replicate(1000, haveCommon())) / 1:1000, type = "l")
lines(cumsum(replicate(1000, haveCommon())) / 1:1000, col = "red")
lines(cumsum(replicate(1000, haveCommon())) / 1:1000, col = "blue")
```
\
## A more serious example: climate change {#climate}
```{r,echo=FALSE}
globalTemp <- read.csv("data/annual_temp.csv")
kable(globalTemp[1:5], "html", table.attr='id="dt_global_temp"')
```
## Change in temperature (global average deviation) since 1851
```{r}
library(lattice)
xyplot(Temp ~ Year, data = globalTemp, grid = TRUE)
```
\
## Change in atmospheric carbon dioxide
```{r}
xyplot(CO2 ~ Year, data = globalTemp, grid = TRUE)
```
\
## Does change in $CO_2$ explain temperature rise?
```{r}
xyplot(Temp ~ CO2, data = globalTemp, grid = TRUE, type = c("p", "r")) # include OLS regression line
```
\
## Fitting the regression model
```{r}
fm = lm(Temp ~ 1 + CO2, data = globalTemp)
coef(fm) # estimated regression coefficients
```
. . .
We can confirm using a general optimizer:
```{r}
SSE = function(beta)
{
with(globalTemp,
sum((Temp - beta[1] - beta[2] * CO2)^2))
}
optim(c(0, 0), fn = SSE)
```
## Fitting the regression model
* `lm()` gives exact solution and more statistically relevant details
```{r}
summary(fm)
```
## Changing the model-fitting criteria
- Suppose we wanted to minimize _sum of absolute errors_ instead of sum of squares
- No closed form solution any more, but general optimizer will still work:
```{r}
SAE = function(beta)
{
with(globalTemp,
sum(abs(Temp - beta[1] - beta[2] * CO2)))
}
opt = optim(c(0, 0), fn = SAE)
opt
```
## Changing the model-fitting criteria
- Compare with least squares line
```{r}
coef(fm) # least squared errors
opt$par # least absolute errors
```
\
* The two lines are virtually identical in this case
* This is not always true
## Another example: number of phone calls per year in Belgium
```{r}
data(phones, package = "MASS")
xyplot(calls ~ year, data = phones, grid = TRUE)
```
\
## Another example: number of phone calls per year in Belgium
```{r}
fm2 <- lm(calls ~ year, data = phones)
SAE = function(beta)
{
with(phones,
sum(abs(calls - beta[1] - beta[2] * year)))
}
opt = optim(c(0, 0), fn = SAE)
```
. . .
```{r}
coef(fm2) # least squared errors
opt$par # least absolute errors
```
* The two lines are quite different
* The second line is an example of _robust regression_
## Another example: number of phone calls per year in Belgium
```{r fig.height=7}
xyplot(calls ~ year, data = phones, grid = TRUE,
panel = function(x, y, ...) {
panel.xyplot(x, y, ...)
panel.abline(fm2, col = "red") # least squared errors
panel.abline(opt$par, col = "blue") # least absolute errors
})
```
\
## Summary
* Conventional statistical learning focuses on problems that can be "solved" analytically
. . .
* Numerical solutions are also valid solutions... but potentially difficult to obtain
* R makes it _easy_ to obtain numerical solutions and compare with traditional solutions
* We will come back to this idea when we next discuss the origins of R
# A very brief history of R
## What is R?
From its own website:
> R is a free software environment for statistical computing and graphics.
. . .
> It is a GNU project which is similar to the S language and
> environment which was developed at Bell Laboratories (formerly AT&T,
> now Lucent Technologies) by John Chambers and colleagues. R can be
> considered as a different implementation of S.
## The origins of S
- Developed at Bell Labs (statistics research department) 1970s onwards
- Primary goals
- Interactivity: Exploratory Data Analysis vs batch mode
- Flexibility: Novel vs routine methodology
- Practical: For actual use, not (just) academic research
. . .
John Chambers received the prestigious _ACM Software System Award_ in 1998
> For The S system, which has forever altered how people analyze,
> visualize, and manipulate data.
## The origins of R
- Early 1990s: Started as teaching tool by Robert Gentleman & Ross
Ihaka at the University of Auckland
- 1995: Convinced by Martin Mächler to release as Free Software (GPL)
- 2000: Version 1.0 released
Has since far surpassed S in popularity
## Number of R packages on CRAN
```{r, echo=FALSE,fig.width=11,fig.height=9}
cranpkgs <- read.csv("cran-packages.csv", skip = 1, stringsAsFactors = FALSE)
cranpkgs$date <- as.Date(cranpkgs$date, "%Y-%m-%d")
xyplot(pkgs ~ date, cranpkgs, grid = TRUE, aspect = "xy")
```
\
## Why the success? The user's perspective {#user}
- R is designed for data analysis
- Basic data structures are vectors
- Large collection of statistical functions
- Advanced statistical graphics capabilities
- The vast majority of R users use it as a statistical toolbox
- R "base" comes with a large suite of statistical modeling and graphics functions
- If these are not enough, more than 10000 add-on packages are available
## The developer's perspective
- Easy dissemination of research (through add-on packages)
- Rapid prototyping
- Interfaces to external software
## Rapid prototyping
John Chambers, _Programming with Data_:
> S is a programming language and environment for all kinds of
> computing involving data. It has a simple goal: _To turn ideas into
> software, quickly and faithfully._
. . .
A silly example: generate Fibonacci sequence
```{r }
fibonacci <- function(n) {
if (n < 2)
x <- seq(length = n) - 1
else {
x <- c(0, 1)
while (length(x) < n) {
x <- c(x, sum(tail(x, 2)))
}
}
x
}
fib10 <- fibonacci(10)
fib10
```
## Also easy to call C for efficiency
```{r, echo=FALSE}
cprog <- "
#include
SEXP fibonacci_c(SEXP nr)
{
int i, n = INTEGER_VALUE(nr);
SEXP ans = PROTECT(NEW_INTEGER(n));
int *x = INTEGER_POINTER(ans);
x[0] = 0; x[1] = 1;
for (i = 2; i < n; i++) x[i] = x[i-1] + x[i-2];
UNPROTECT(1);
return ans;
}
"
cat(cprog, file = "fib.c")
```
File `fib.c`:
```c
#include
SEXP fibonacci_c(SEXP nr)
{
int i, n = INTEGER_VALUE(nr);
SEXP ans = PROTECT(NEW_INTEGER(n));
int *x = INTEGER_POINTER(ans);
x[0] = 0; x[1] = 1;
for (i = 2; i < n; i++) x[i] = x[i-1] + x[i-2];
UNPROTECT(1);
return ans;
}
```
Compile into shared library:
```
$ R CMD SHLIB fib.c
```
```{r,echo=FALSE,results="hide"}
cat(system("R CMD SHLIB fib.c", intern = TRUE)[c(3, 4)], sep = "\n")
```
Load into R and call:
```{r}
dyn.load("fib.so")
cfib10 = .Call("fibonacci_c", as.integer(10))
cfib10
```
## Even easier to call C++ with Rcpp package
```{r, echo=FALSE}
cppprog <- "
#include
using namespace Rcpp;
// [[Rcpp::export]]
NumericVector fibonacci_cpp(int n)
{
NumericVector x(n);
x[0] = 0; x[1] = 1;
for (int i = 2; i < n; i++) x[i] = x[i-1] + x[i-2];
return x;
}
"
cat(cppprog, file = "fib.cpp")
```
File `fib.cpp`:
```c++
#include
using namespace Rcpp;
// [[Rcpp::export]]
NumericVector fibonacci_cpp(int n)
{
NumericVector x(n);
x[0] = 0; x[1] = 1;
for (int i = 2; i < n; i++) x[i] = x[i-1] + x[i-2];
return x;
}
```
Compile and call:
```{r}
Rcpp::sourceCpp("fib.cpp")
fibonacci_cpp(10)
```
## Rapid prototyping: flexibility and extensibility
- Powerful built-in tools
- Programming language
- Compiled code for efficiency
## Another strength: Interfaces
- Not all useful software developed by R community
- Core open source philosophy: code re-use
- Creating interfaces with external software is relatively easy
- Example: Keras / TensorFlow
## Keras {#keras}
- Deep learning framework based on TensorFlow
- R interface through package [keras](https://blog.rstudio.com/2017/09/05/keras-for-r/)
## Example: classify handwritten digits
```{r}
library(keras)
mnist <- dataset_mnist()
x_train <- mnist$train$x # each sample is a 28x28 grayscale image
y_train <- mnist$train$y # correct classification (0,1,2,...,9)
x_test <- mnist$test$x
y_test <- mnist$test$y
```
. . .
```{r,fig.width=20,fig.height=4}
xtrain.100 <- as.data.frame.table(x_train[1:100,,])
levelplot(Freq ~ Var3 + Var2 | Var1, data = xtrain.100, strip = FALSE, scales = list(draw = FALSE),
ylim = c(28, 1), colorkey = FALSE, col.regions = rev(grey.colors(20)), xlab = NULL, ylab = NULL, aspect = 1)
```
\
## Transform data
- Reshape data (to vector) and rescale
```{r}
# reshape each 28x28 image matrix to 784-vector
dim(x_train) <- c(nrow(x_train), 784)
dim(x_test) <- c(nrow(x_test), 784)
# rescale grayscale values (0-225) to (0,1)
x_train <- x_train / 255
x_test <- x_test / 255
y_train <- to_categorical(y_train, 10)
y_test <- to_categorical(y_test, 10)
```
## Define model
- A Keras model is a way to organize layers
- Define a sequential model (a linear stack of layers)
```{r}
model <- keras_model_sequential()
layer_dense(model, units = 256, activation = "relu", input_shape = c(784))
layer_dropout(model, rate = 0.4)
layer_dense(model, units = 128, activation = "relu")
layer_dropout(model, rate = 0.3)
layer_dense(model, units = 10, activation = "softmax")
summary(model)
```
## Compile and train model
```{r}
compile(model,
loss = "categorical_crossentropy",
optimizer = optimizer_rmsprop(),
metrics = c("accuracy"))
history <- fit(model,
x_train, y_train,
epochs = 30, batch_size = 128,
validation_split = 0.2)
```
## Evaluate model
```{r}
p <- plot(history)
p
```
\
## Results on test data
```{r}
pred_class <- predict_classes(model, x_test)
pred_class[1:20]
y_test[1:20,]
```
## Misclassification rate in test data
```{r}
ctab <- table(pred_class, apply(y_test, 1, which.max)-1)
ctab
sum(diag(ctab)) / sum(ctab)
```
## Another interface: plotly {#plotly}
- Plotly: a Javascript library for visualization
- R interface provided by the `plotly` R package
```{r,eval=FALSE}
library(plotly)
ggplotly(p)
```
## More HTML-based applications
- [DataTable (plug-in for jQuery)](https://datatables.net/) - [example earlier](#climate)
- [HTML widgets](http://www.htmlwidgets.org/)
- [Shiny Apps](https://shiny.rstudio.com/)
## Parting comments: reproducible documents
- Creating reports / presentations with numerical analysis is usually a two-step process:
- Do the analysis using a computational software
- Write report in a word processor, copy-pasting results
- R makes it very convenient to write "literate documents" that
contain both analsyis code and report text
- Basic idea:
- Start with source text file containing code+text
- Transform file by running code and embedding results
- Produces another text file (LaTeX, HTML, markdown)
- Processed further using standard tools
- Example: this presentation is created from
[this source file](roverview.rmd) (R Markdown) using
[knitr](https://yihui.name/knitr/) and [pandoc](http://pandoc.org/)
- As the source format is markdown, output could also be
[PDF](roverview.pdf) instead of HTML