Visualizing Large Data: Broad Themes
Deepayan Sarkar
Big data
The data we typically work with is rectangular / spreadsheet \(X_{n \times p}\)
The \(n\) rows are observations (units)
The \(p\) columns are variables (measurements) on each unit
When do we consider such data to “big data” or “large data”?
Big data can mean several things
Many interesting statistical questions arise when we have
Many observations (\(n\) large)
Many variables (\(p\) large)
But can also lead to engineering issues
Engineering / informatics issues
Lack of structure
Raw data may not have \(X_{n \times p}\) structure
Genomic expression, gene sequencing
Satellite / medical imaging, video
May need preprocessing before analysis
May or may not need statistical methodology
Visualization
Usually has two main purposes:
Exploration: Intermediate steps as part of interactive analysis
Presentation: Summarize or report findings for external audience
In both cases, the data are usually summarized or preprocessed before being visualized
This may still happen dynamically / on demand using a distributed cloud-based system
Example: NASA Wordview (Satellite data visualization)
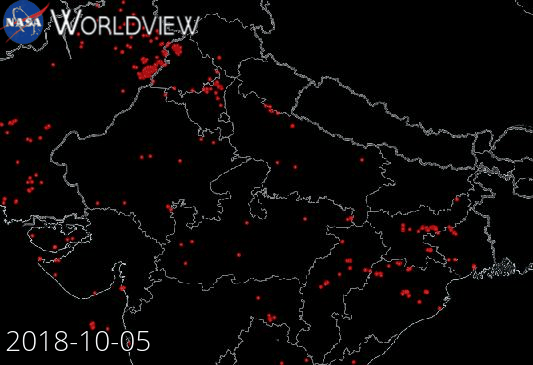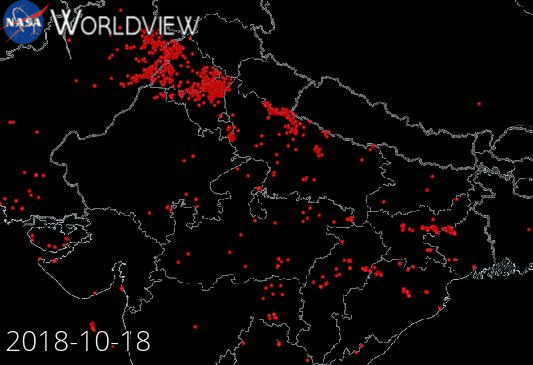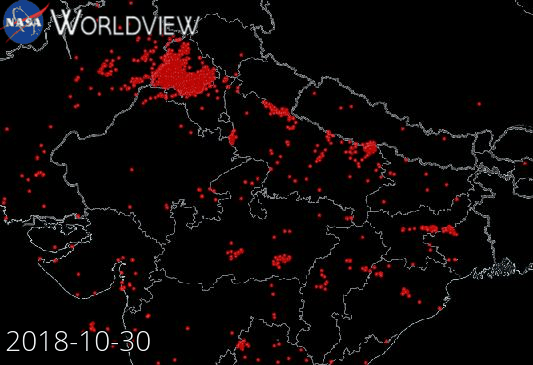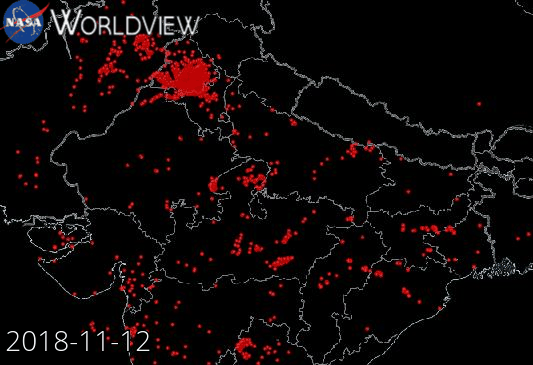
Our starting point
We will assume data has been suitably summarized
We will also assume it has the standard \(X_{n \times p}\) format
\(n\) can be moderately large (but still fits in memory)
\(p\) is usually limited for a particular visualization
Example datasets: airquality (small \(n\))
'data.frame': 153 obs. of 6 variables:
$ Ozone : int 41 36 12 18 NA 28 23 19 8 NA ...
$ Solar.R: int 190 118 149 313 NA NA 299 99 19 194 ...
$ Wind : num 7.4 8 12.6 11.5 14.3 14.9 8.6 13.8 20.1 8.6 ...
$ Temp : int 67 72 74 62 56 66 65 59 61 69 ...
$ Month : int 5 5 5 5 5 5 5 5 5 5 ...
$ Day : int 1 2 3 4 5 6 7 8 9 10 ... Ozone Solar.R Wind Temp Month Day
1 41 190 7.4 67 5 1
2 36 118 8.0 72 5 2
3 12 149 12.6 74 5 3
4 18 313 11.5 62 5 4
5 NA NA 14.3 56 5 5
6 28 NA 14.9 66 5 6
7 23 299 8.6 65 5 7
8 19 99 13.8 59 5 8
9 8 19 20.1 61 5 9
10 NA 194 8.6 69 5 10
11 7 NA 6.9 74 5 11
12 16 256 9.7 69 5 12
13 11 290 9.2 66 5 13
14 14 274 10.9 68 5 14
15 18 65 13.2 58 5 15Example datasets: gapminder (moderate \(n\))
# A tibble: 1,704 x 6
country continent year lifeExp pop gdpPercap
<fct> <fct> <int> <dbl> <int> <dbl>
1 Afghanistan Asia 1952 28.8 8425333 779.
2 Afghanistan Asia 1957 30.3 9240934 821.
3 Afghanistan Asia 1962 32.0 10267083 853.
4 Afghanistan Asia 1967 34.0 11537966 836.
5 Afghanistan Asia 1972 36.1 13079460 740.
6 Afghanistan Asia 1977 38.4 14880372 786.
7 Afghanistan Asia 1982 39.9 12881816 978.
8 Afghanistan Asia 1987 40.8 13867957 852.
9 Afghanistan Asia 1992 41.7 16317921 649.
10 Afghanistan Asia 1997 41.8 22227415 635.
# … with 1,694 more rowsExample datasets: vaccines (moderate \(n\))
data(vaccines, package = "highcharter")
vaccines # 'count' is number of measles cases per 100,000 people# A tibble: 3,876 x 3
year state count
<int> <chr> <dbl>
1 1928 Alabama 335.
2 1928 Alaska NA
3 1928 Arizona 201.
4 1928 Arkansas 482.
5 1928 California 69.2
6 1928 Colorado 207.
7 1928 Connecticut 635.
8 1928 Delaware 256.
9 1928 District Of Columbia 536.
10 1928 Florida 120.
# … with 3,866 more rowsExample datasets: NHANES (largish \(n\))
'data.frame': 9575 obs. of 15 variables:
$ Cancer.Incidence: Factor w/ 2 levels "No","Yes": 1 1 1 1 1 1 1 1 1 1 ...
$ Cancer.Death : Factor w/ 2 levels "No","Yes": 1 1 1 1 1 1 1 1 1 1 ...
$ Age : num 46 72 53 49 31 42 27 64 37 42 ...
$ Smoke : Factor w/ 4 levels "Current","Past",..: 3 4 2 4 1 1 1 4 3 2 ...
$ Ed : num 0 0 1 0 0 1 0 0 1 1 ...
$ Race : num 0 0 1 1 1 1 0 0 0 1 ...
$ Weight : num 82.7 85.6 71.5 93.8 81.6 68.3 67.7 86.5 61.7 85 ...
$ BMI : num 29.4 29.6 23.1 30 29.7 21.9 26.3 31.9 20.9 26.7 ...
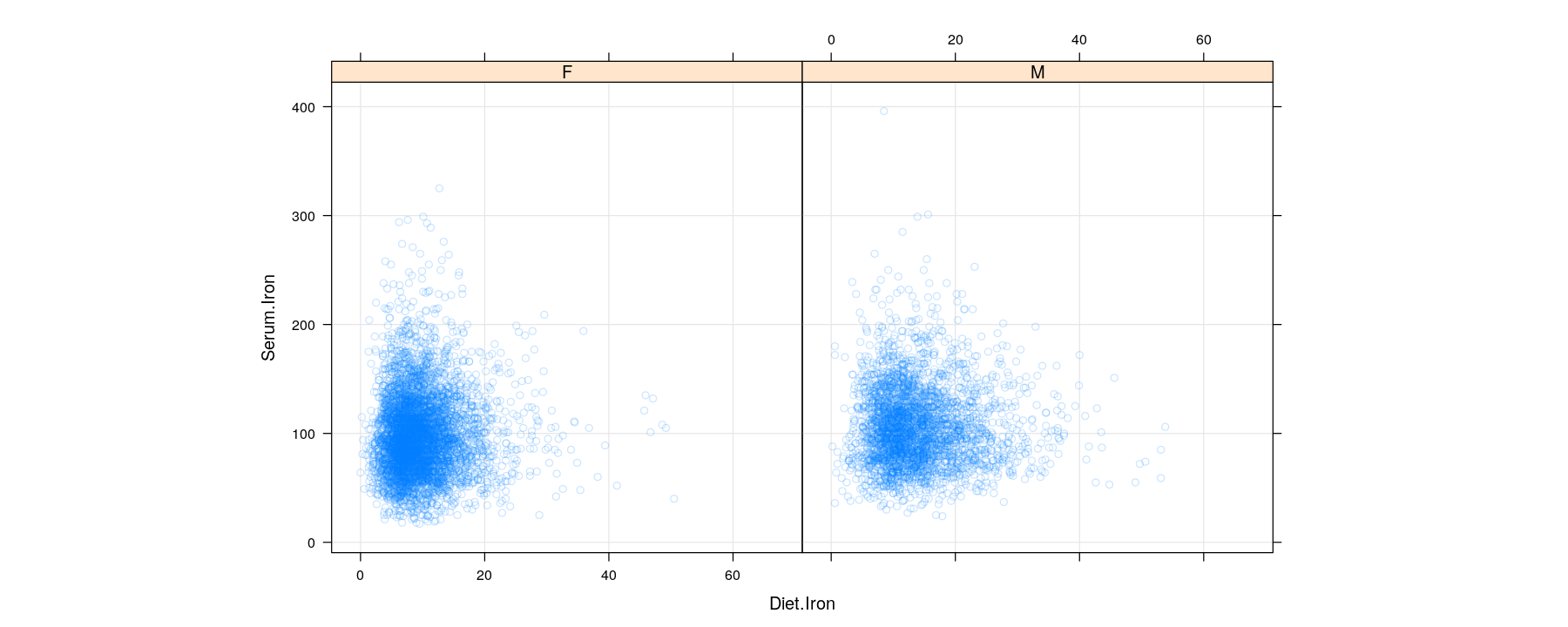
$ Diet.Iron : num 12.5 NA 14.8 24.3 11.8 25.3 25.2 6.8 26.2 11 ...
$ Albumin : num 4.8 4.1 NA 4.3 4.3 4.8 4.3 3.9 4.4 4.7 ...
$ Serum.Iron : num NA NA 135 85 104 124 68 NA 69 89 ...
$ TIBC : num NA NA 334 385 436 285 468 NA 291 456 ...
$ Transferin : num NA NA 40.4 22.1 23.9 43.5 14.5 NA 23.7 19.5 ...
$ Hemoglobin : num 14.7 14.5 16.8 16 16.1 17.1 14.3 15 15.2 16.3 ...
$ Sex : Factor w/ 2 levels "F","M": 2 2 2 2 2 2 2 2 2 2 ...The goal of visualization
Useful visualizations enable us to study relationships
This is usually done through comparison
There are only a few basic objects we usually want to study through visualization
Univariate distributions
Bivariate and trivariate (generally multivariate) relationships
Special case: Relationship with time (time-series) or space (spatial)
Cross-tabulations (counts) summarizing relationships between categorical variables
- We will briefly discuss the common procedures and how they can be adapted for large data
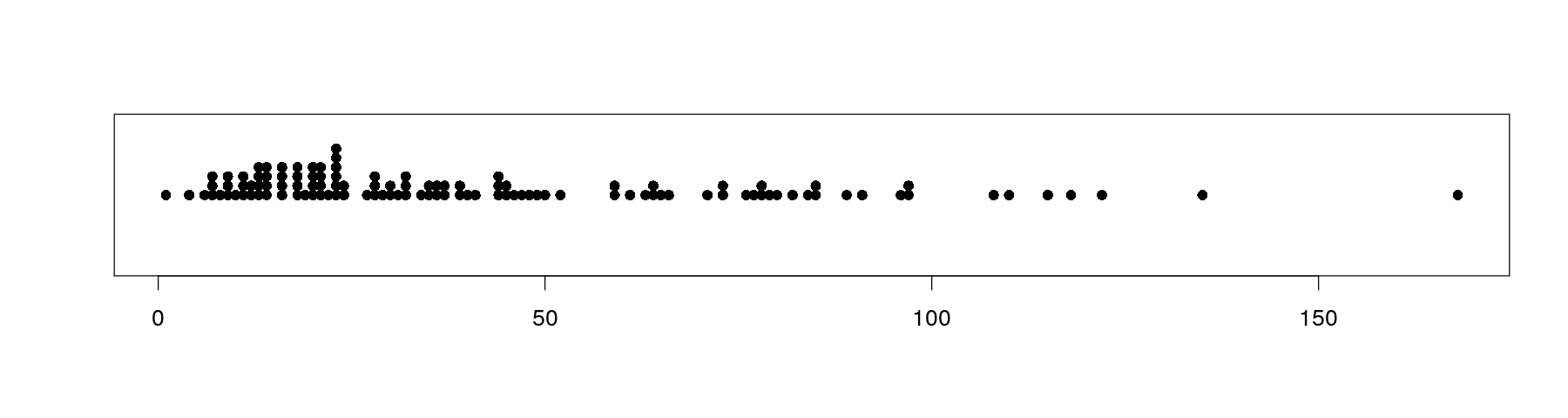
Univariate distributions: strip charts or dot plots
The simplest possible plot of univariate data is to represent each observation by a point
Value of observation is converted into position on x-axis

Univariate distributions: strip charts or dot plots
This overplots multiple data points with same values
A common textbook suggestion is to stack repeated values
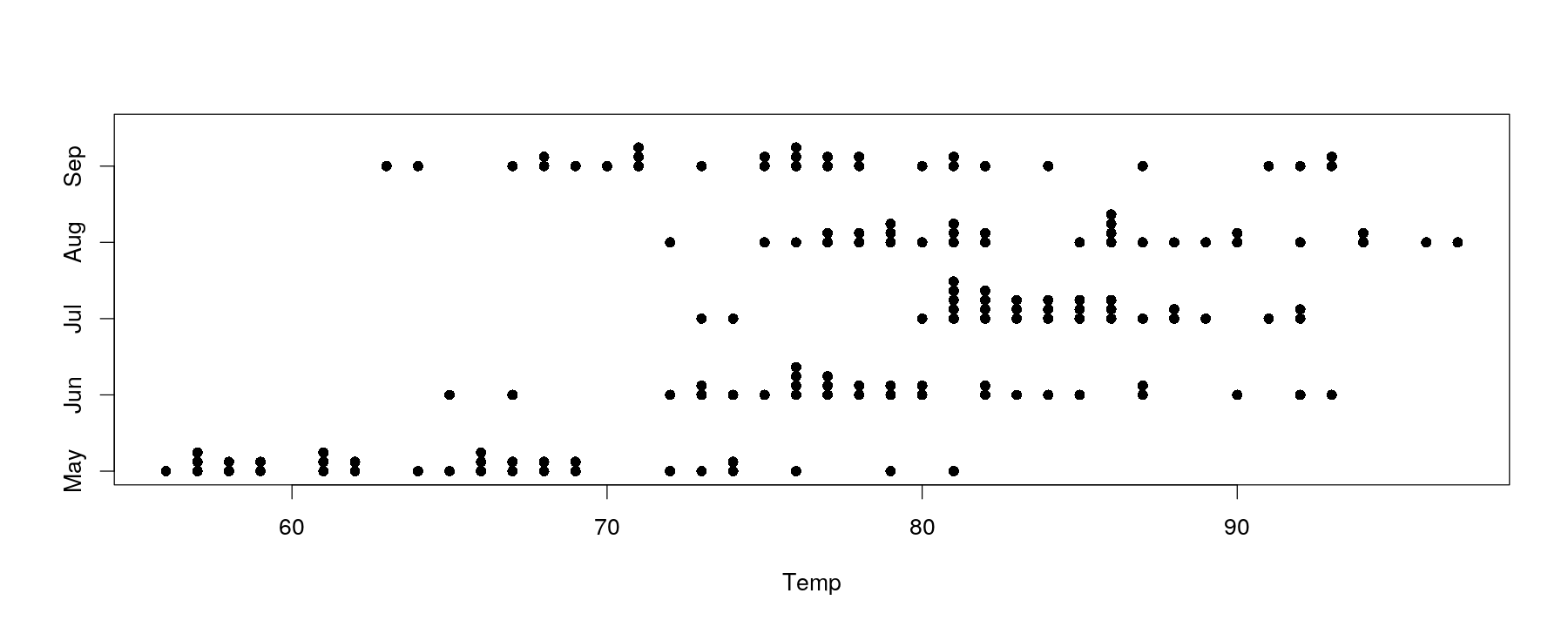
Univariate distributions: comparative strip charts
- Easy to add comparison across categorical variable by encoding as y-coordinate

Univariate distributions: comparative strip charts
- More variation in temperature
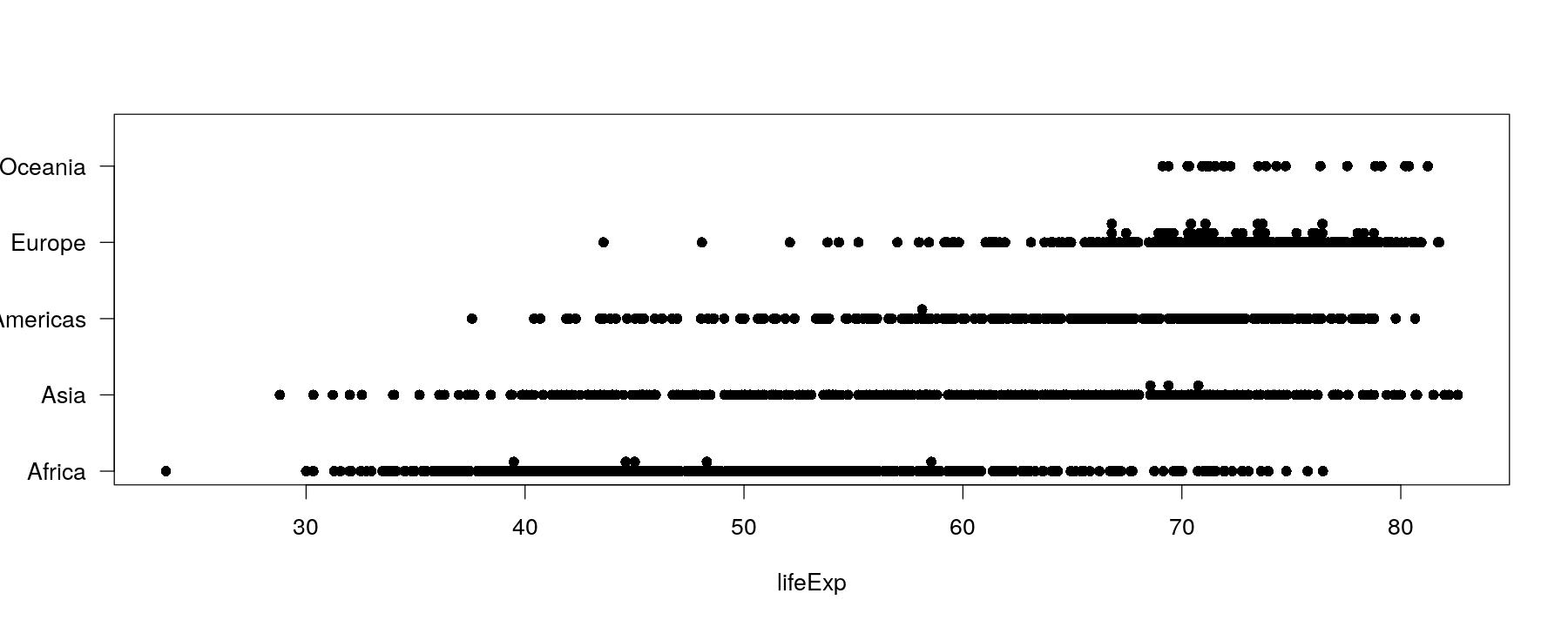
Univariate distributions: comparative strip charts
- This no longer works for moderately sized data (and not many ties)
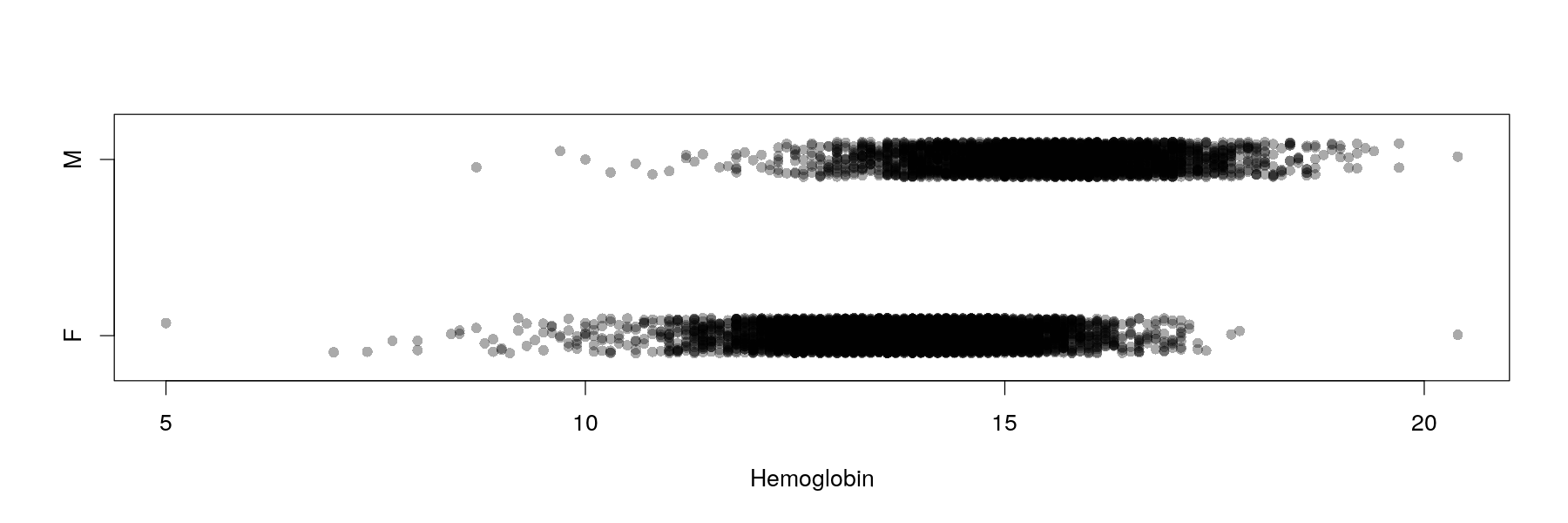
Univariate distributions: comparative strip charts
- Can be improved somewhat by jittering (add random noise) and semi-transparent colors

Univariate distributions: comparative strip charts
- Still not enough when there are too many data points
Univariate distributions: comparative box and whisker plots
Solution: summarize data instead of showing all data points
A common tool is to show quartiles and extremes (and “outliers”)
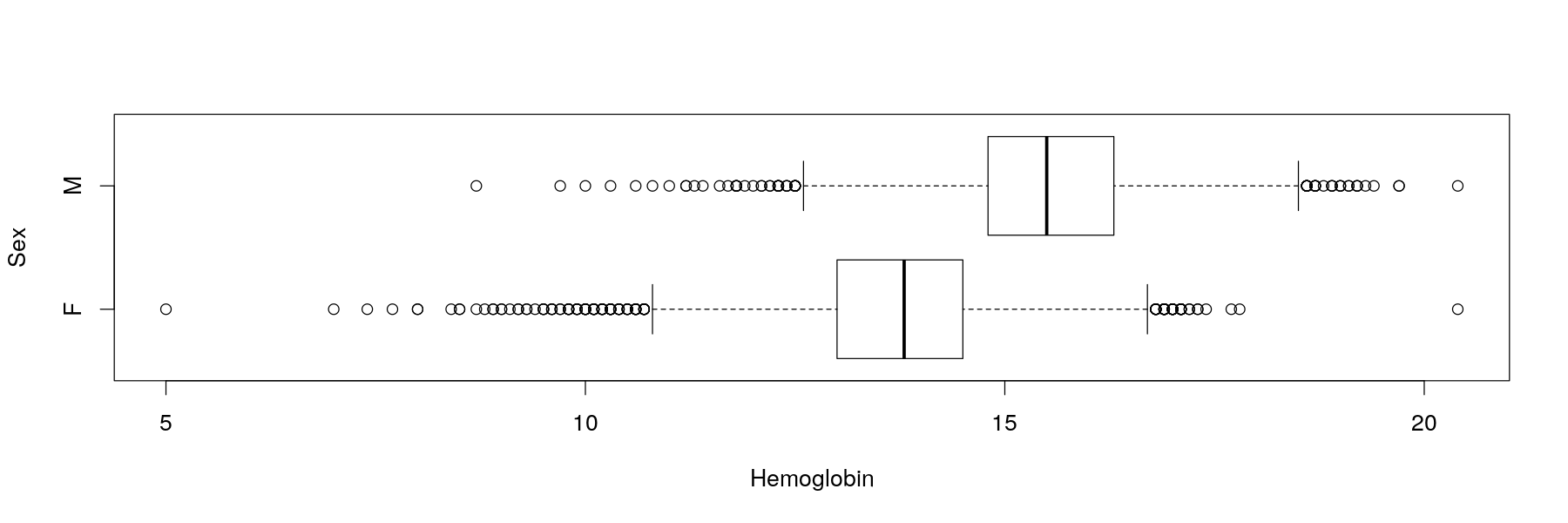
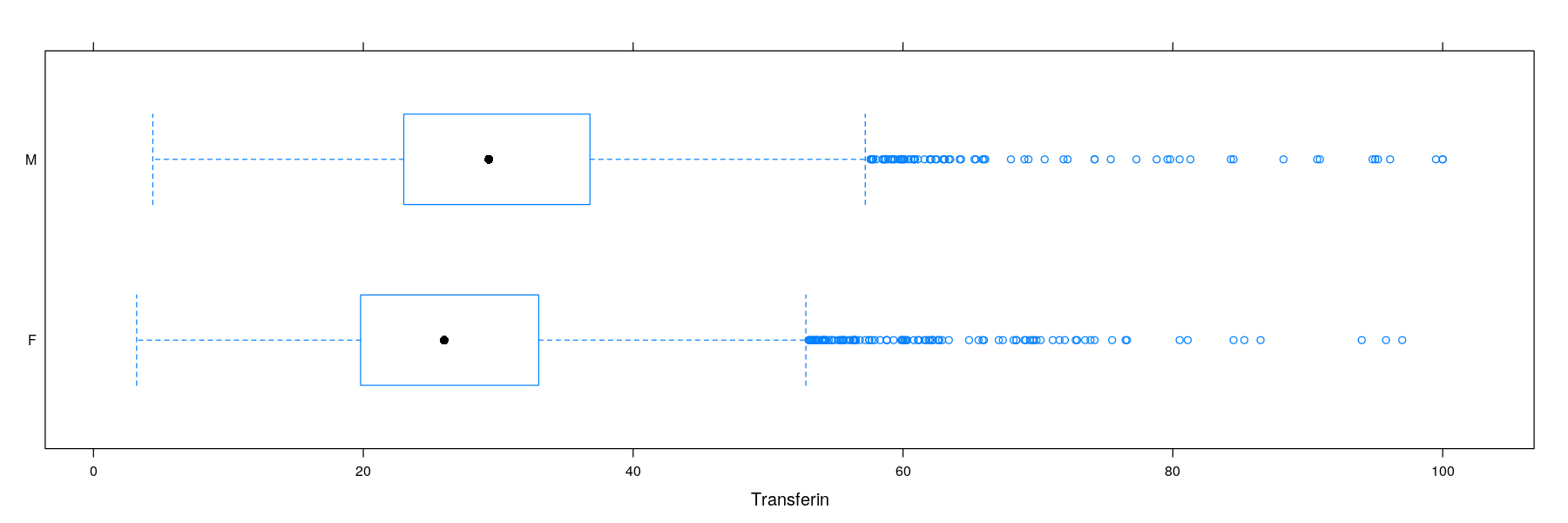
Univariate distributions: comparative box and whisker plots
- A Similar plot using the
latticepackage (more details tomorrow)
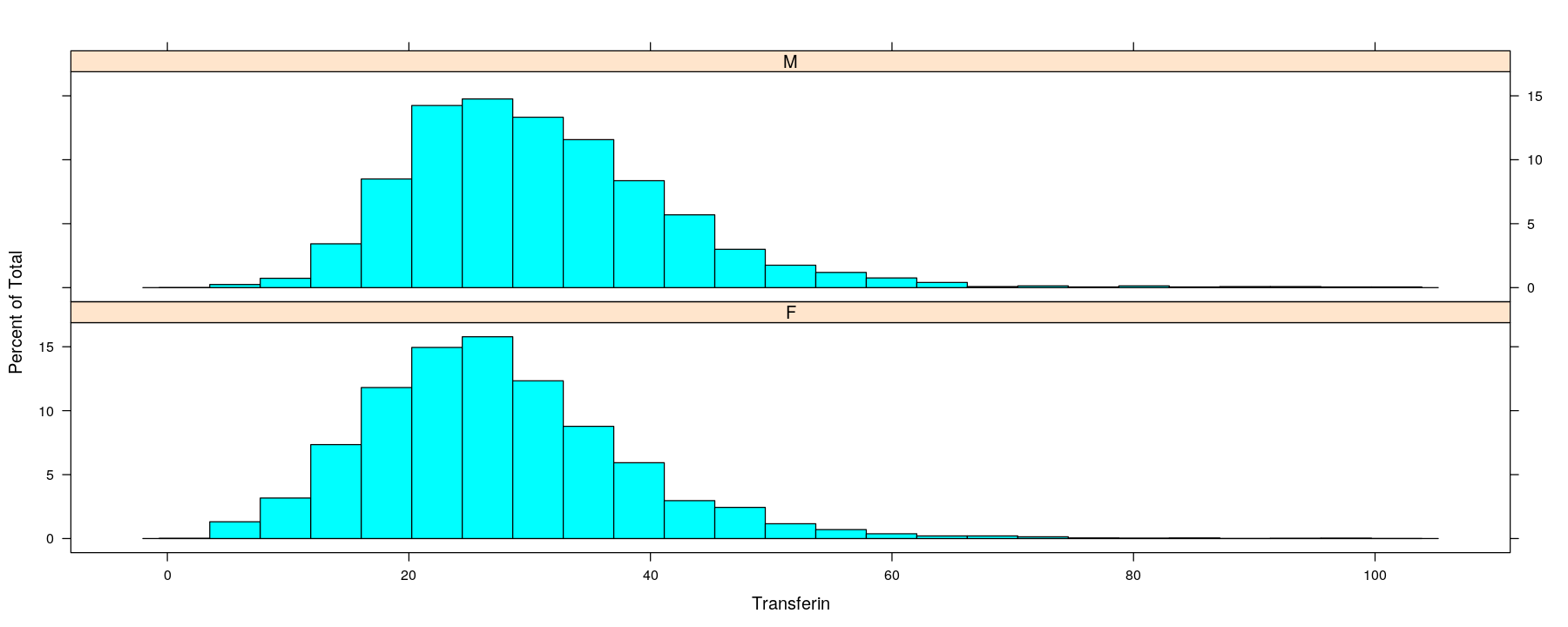
Univariate distributions: comparative histograms
- Another common summary: put data into bins and plot bin counts
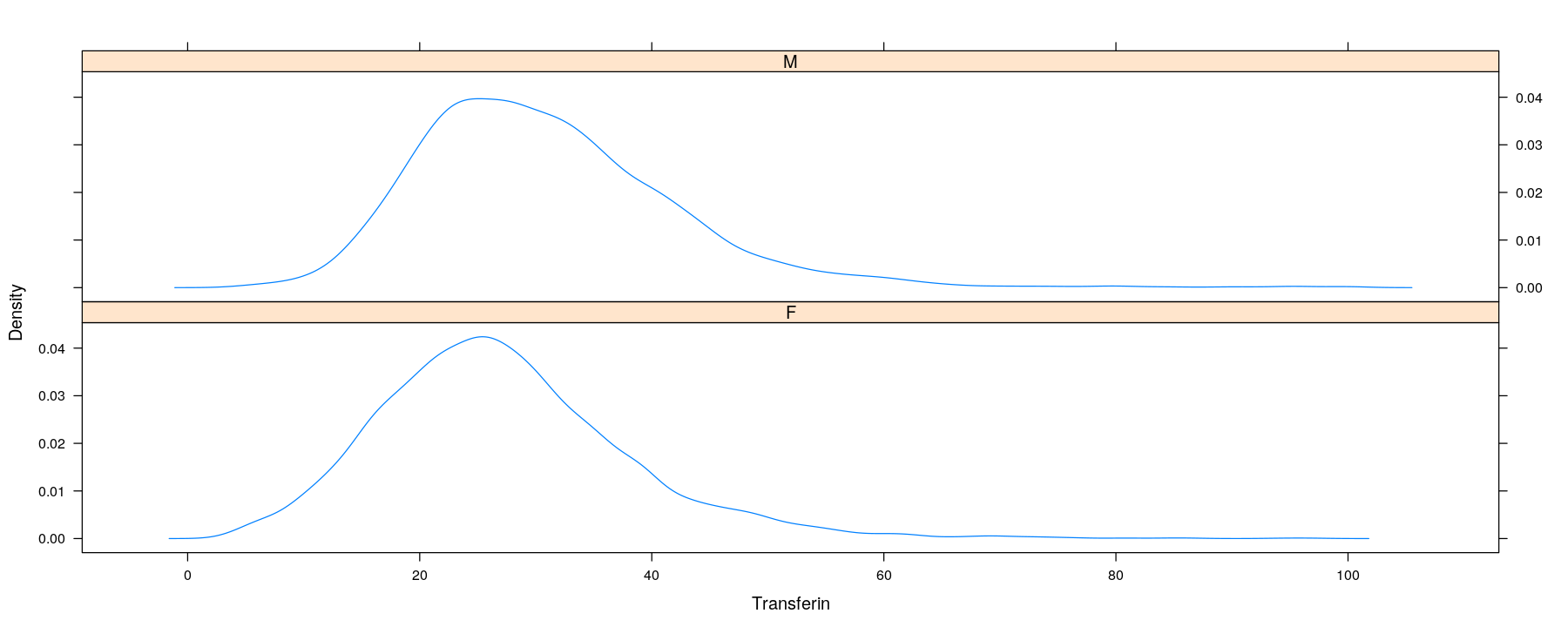
Univariate distributions: kernel density estimates
- A more sophisticated summary: smooth histogram
Univariate distributions: comparative violin plots
- Can be incorporated into the box-and-whisker plot design
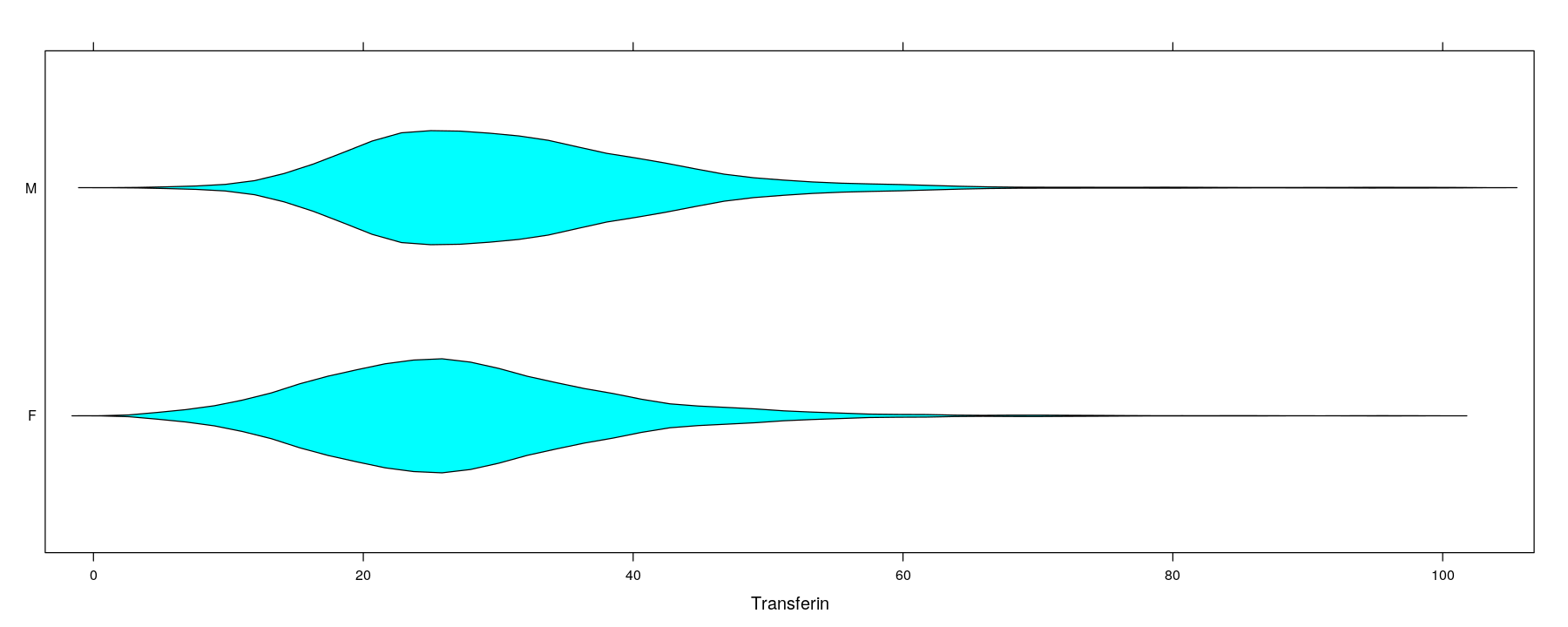
Univariate distributions
Note that all these are essentially generalizations of the basic strip chart
Similar generalizations can be used to study bivariate relationships
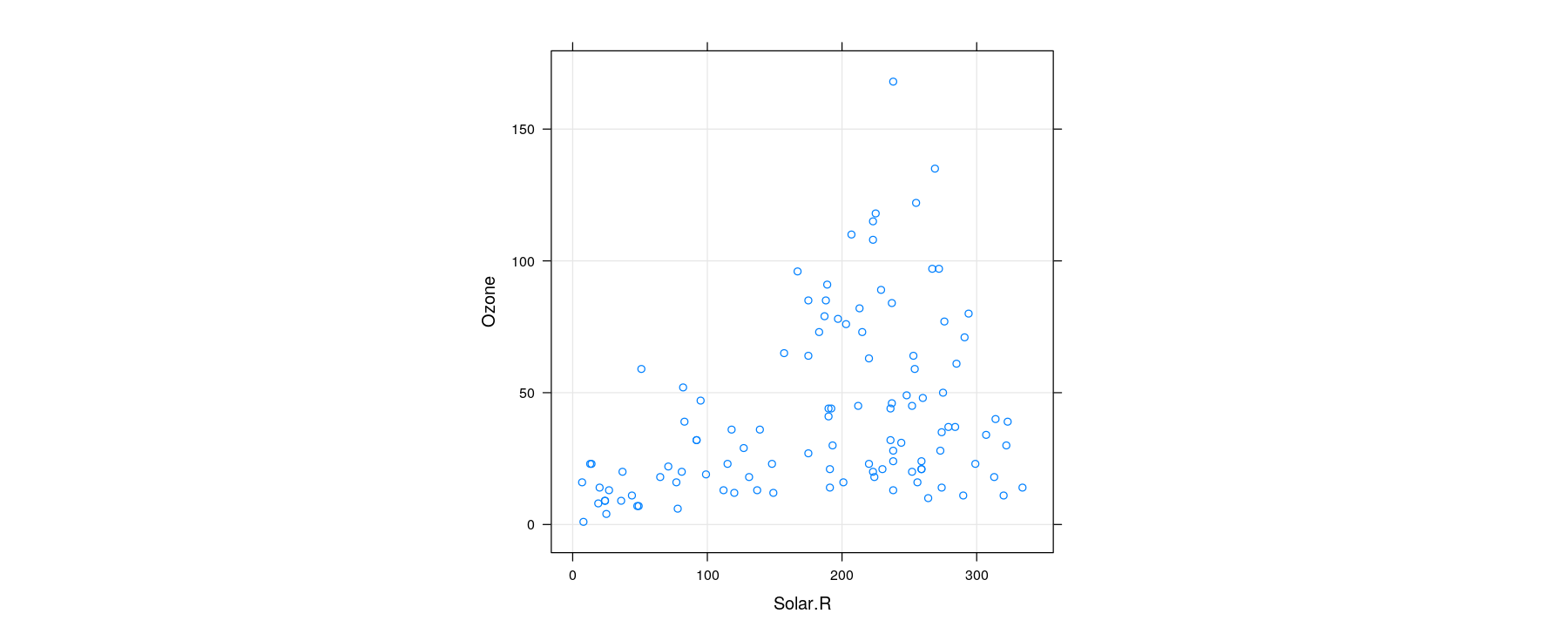
Bivariate distributions: scatter plot
- The basic scatter plot encodes two variables as x- and y-coordinates
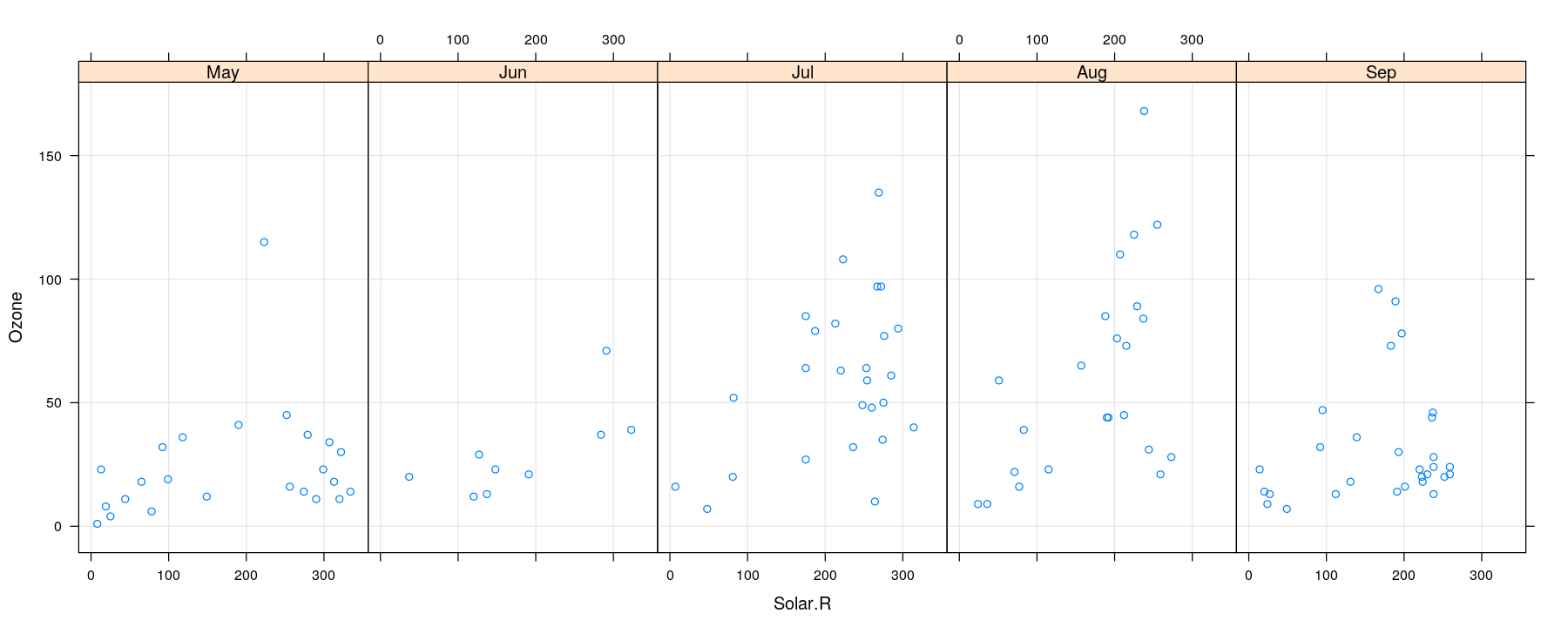
Bivariate distributions: comparative scatter plots
Bivariate distributions: scatter plots using semi-transparent colors
Bivariate distributions: hexagonal binning
Hexagons are preferable over squares for two-dimensional binning (more dense packing)
Bin counts are usually indicated by color (somewhat similar to overplotted semi-transparent points)
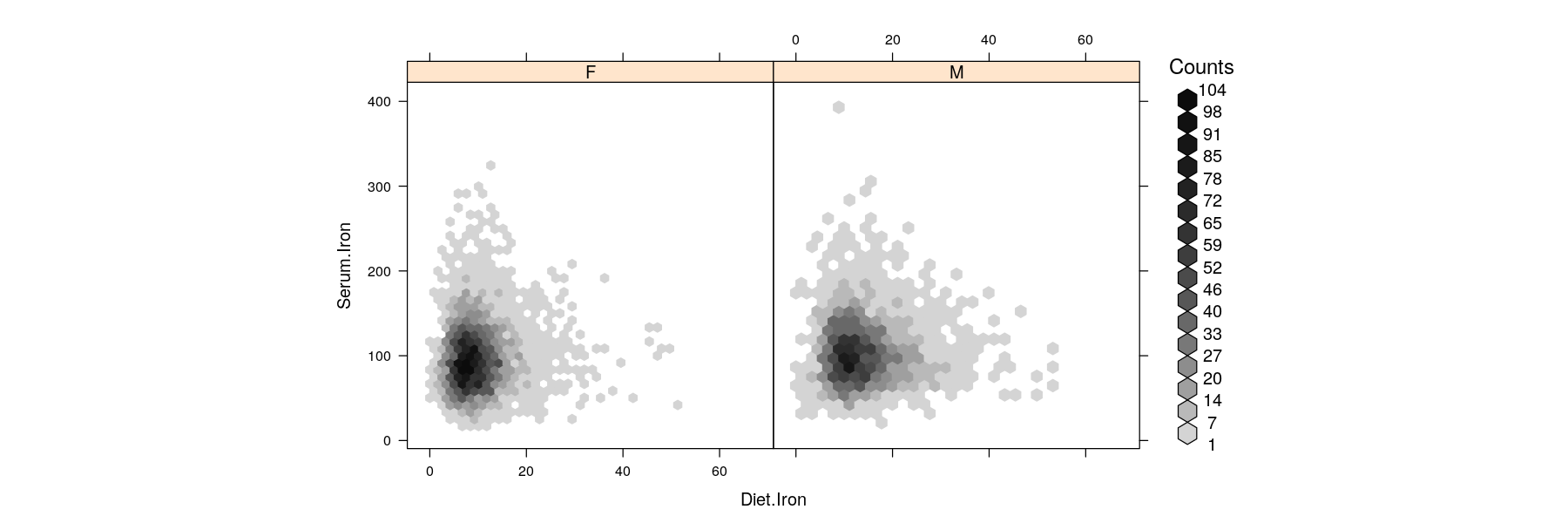
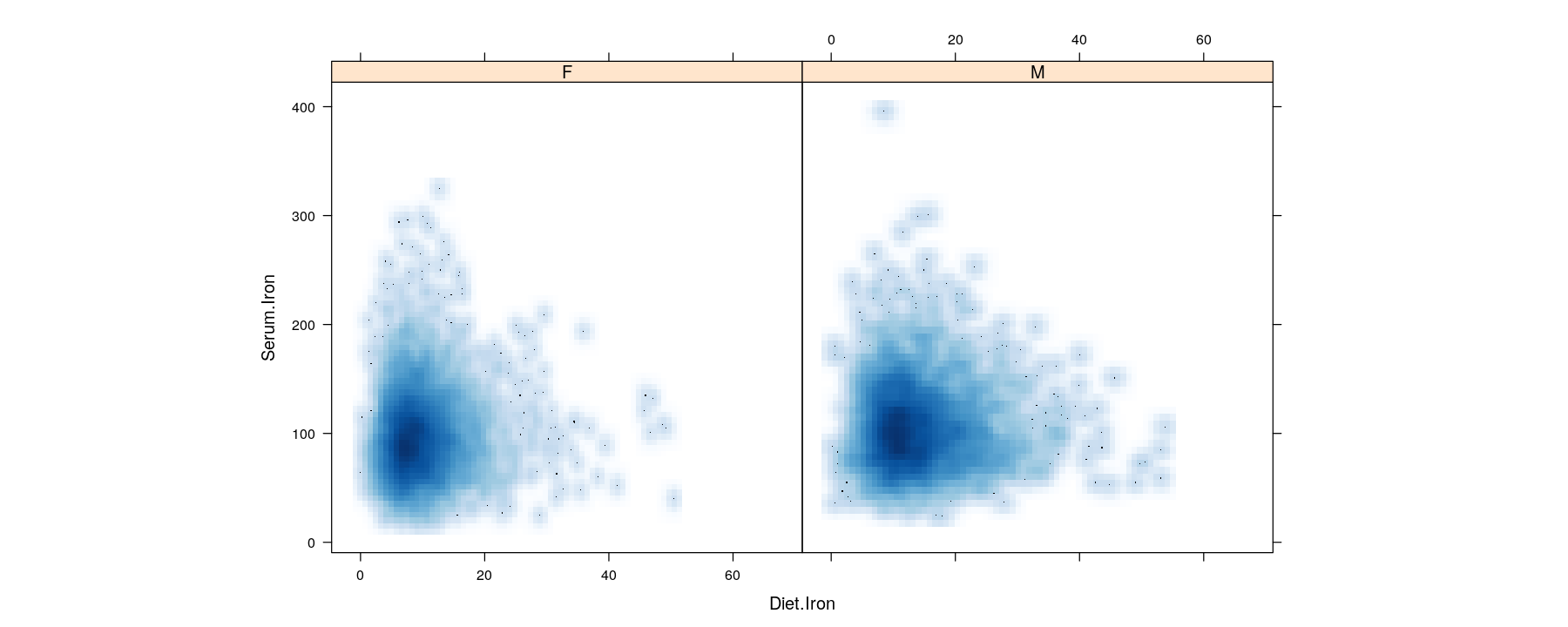
Bivariate distributions: kernel density estimates
- Two-dimensional kernel density estimates can be similarly encoded by color
Tables: Summary measures on categorical attributes
- Small tables are often visualized using bar charts or Cleveland dot plots (summary encoded by position)
dotplot(reorder(country, lifeExp) ~ lifeExp, data = subset(gapminder, continent == "Asia" & year %in% c(1962, 1982, 2002)),
groups = year, auto.key = list(space = "top", columns = 3), par.settings = simpleTheme(pch = 16))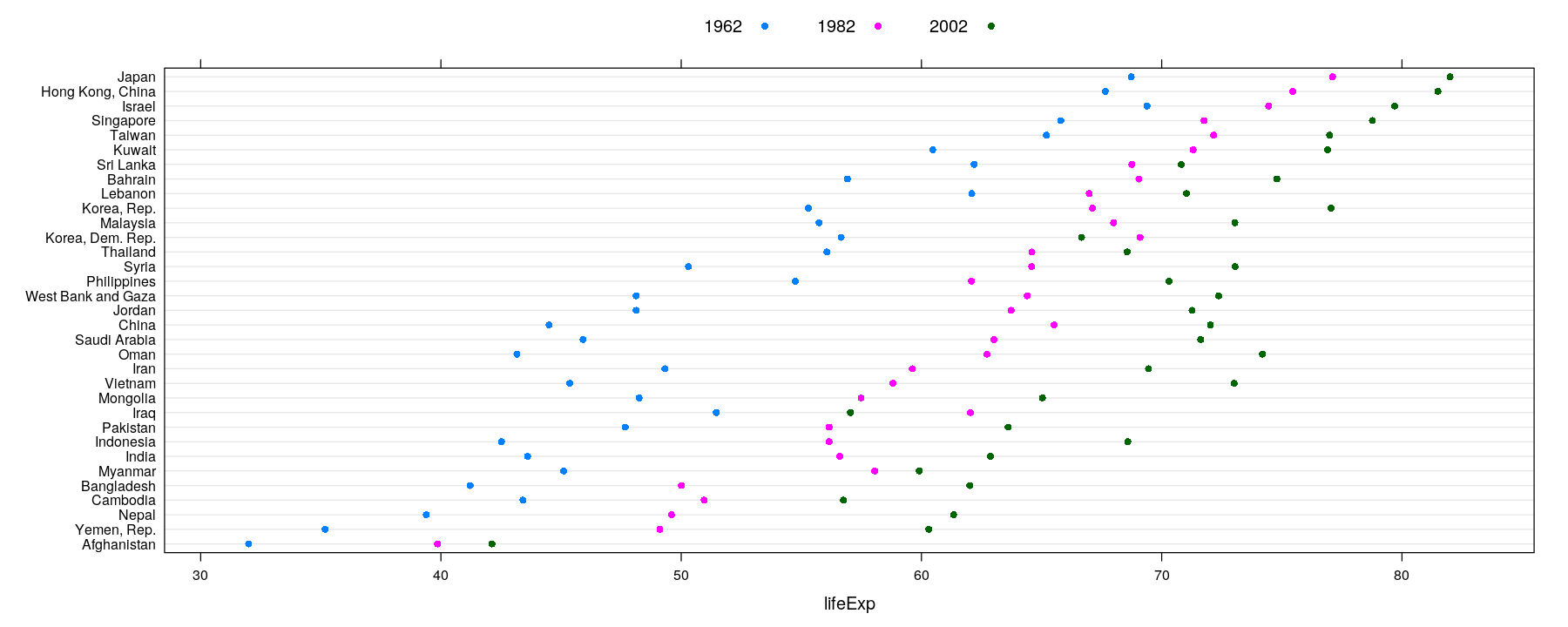
Tables: Summary measures on categorical attributes
- Color can be used to encode value for 2-D tables: e.g., number of measles cases per 100,000 population
levelplot(xtabs(count ~ year + state, data = vaccines), scales = list(x = list(rot = 90)),
col.regions = rev(hcl.colors(16, palette = "Purple-Orange")), aspect = "fill")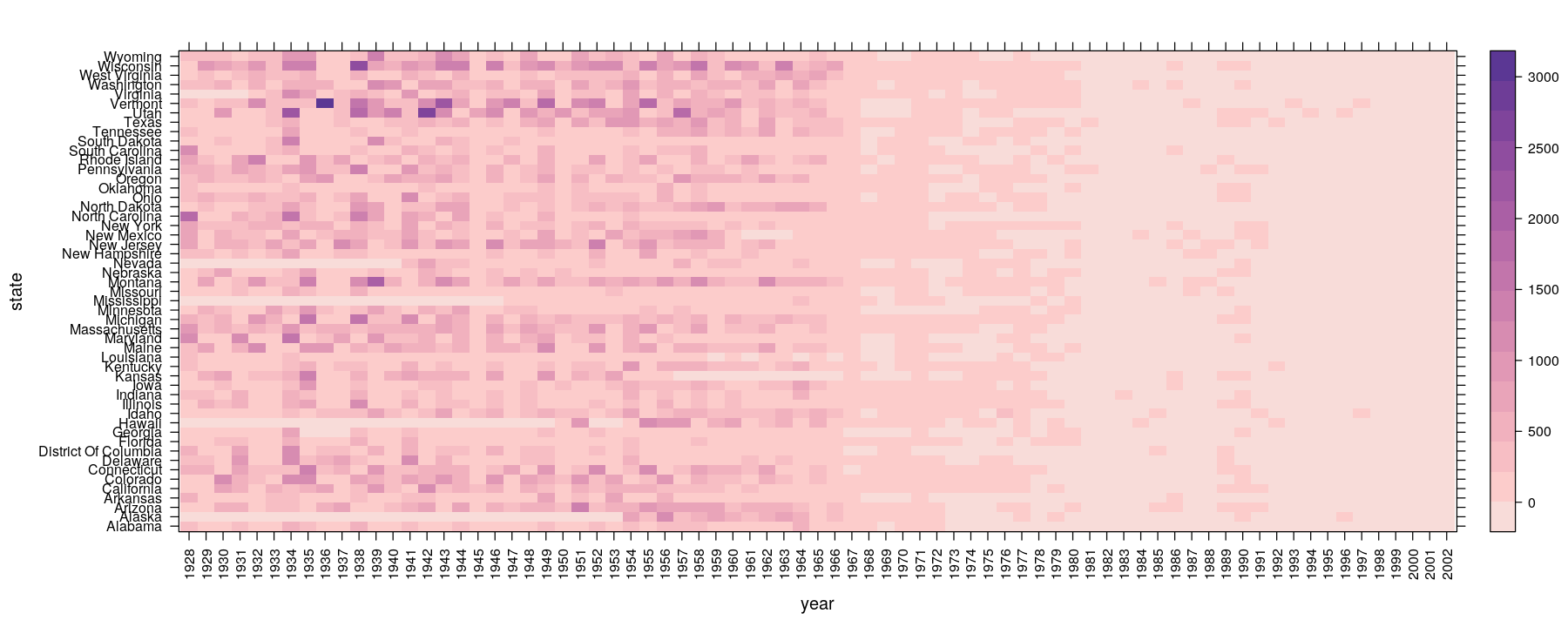
Trivariate data: projection into two-dimensional space
- In principle, three variables can be mapped to x, y, z-coordinates
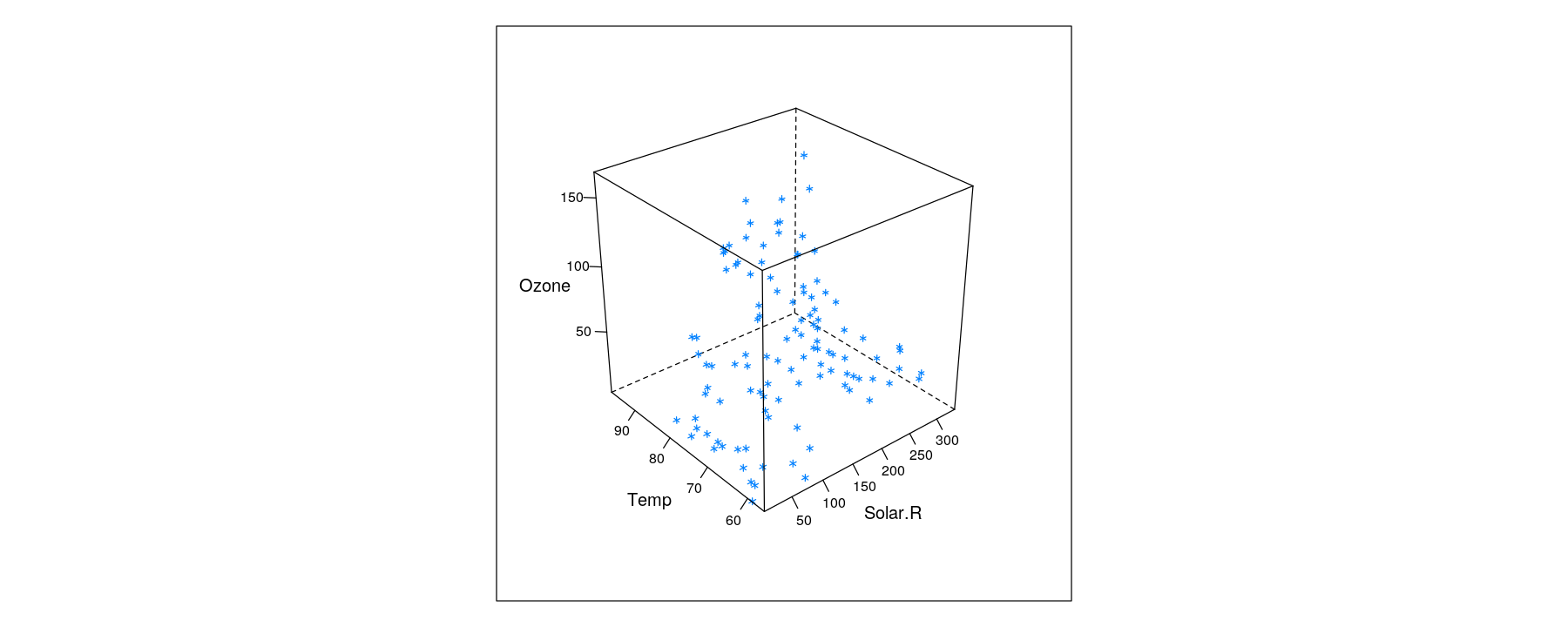
Trivariate data: projection into two-dimensional space
- Interactive version using
rglpackage (interface to OpenGL)
More variables to compare against: conditioning / faceting
- As seen earlier, further categorical variables can be used to compare by superposition
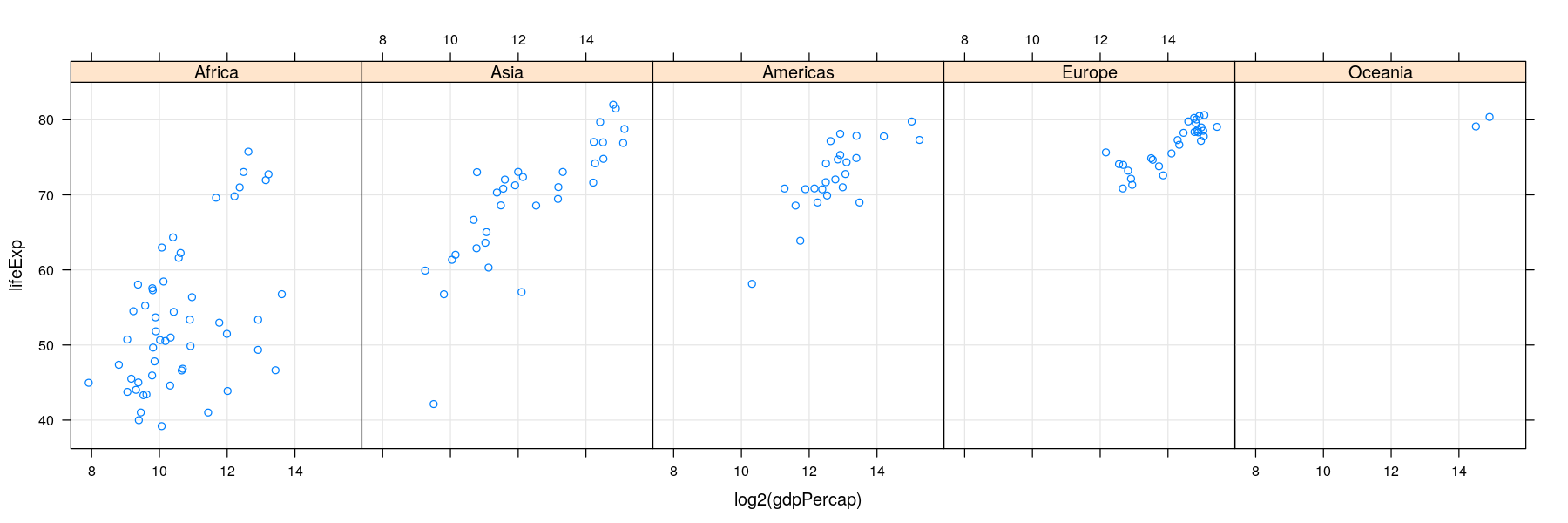
More variables to compare against: conditioning / faceting
- For too many comparisons, a single display page may not be enough
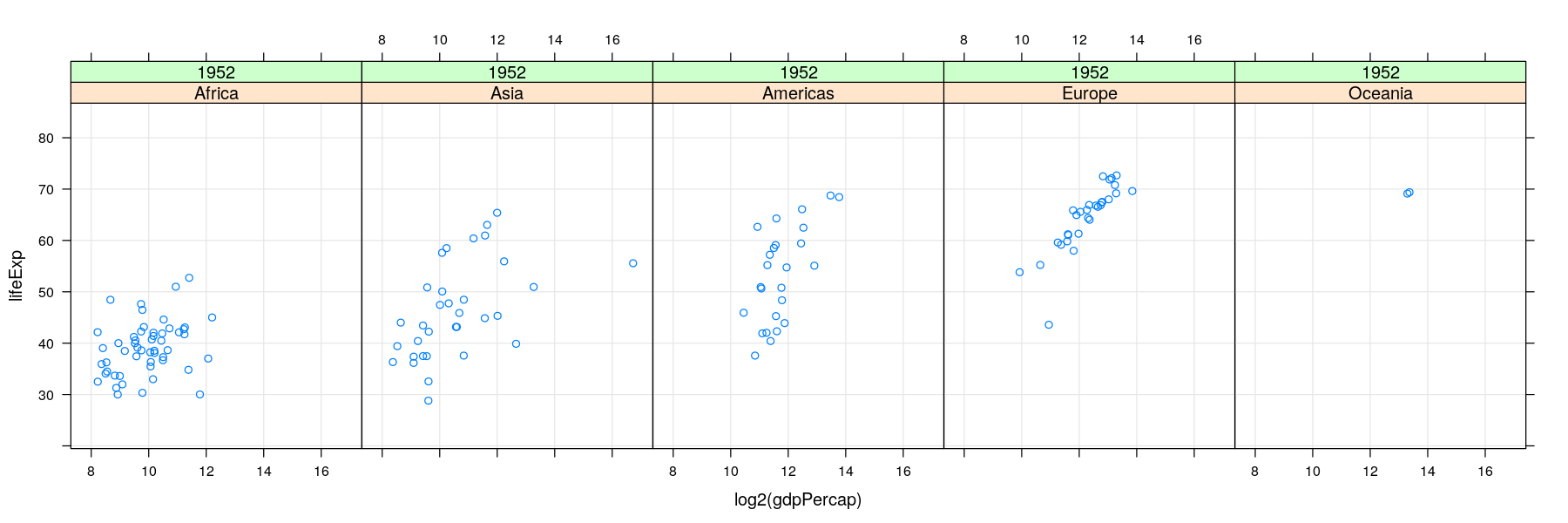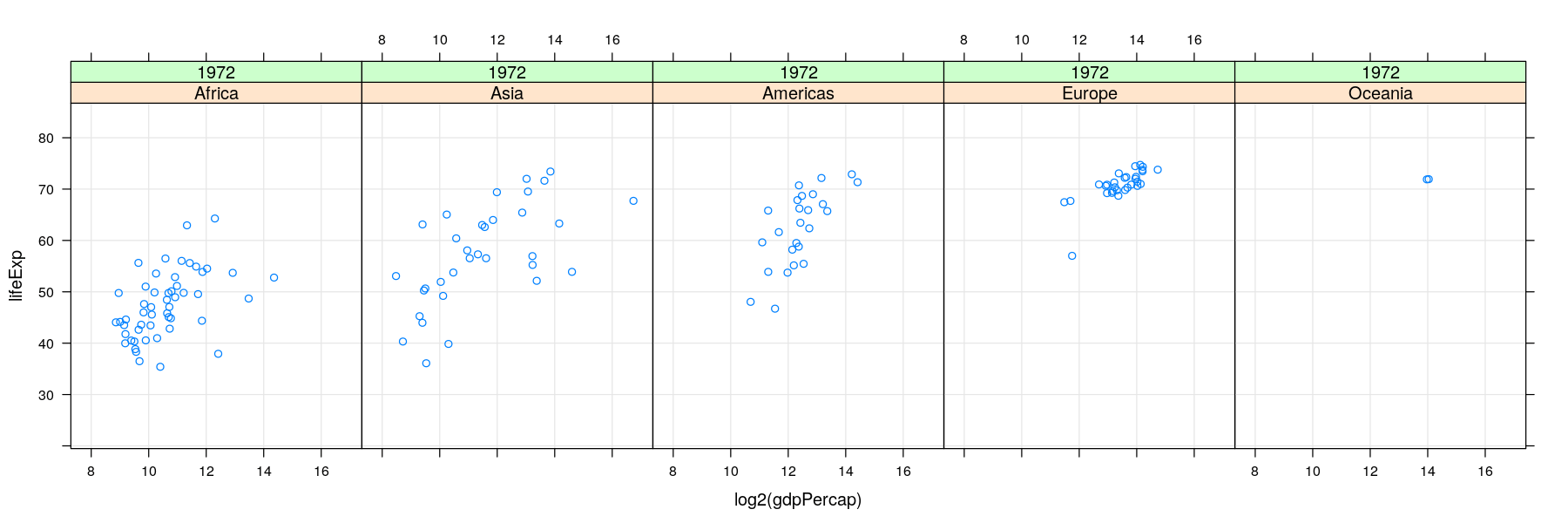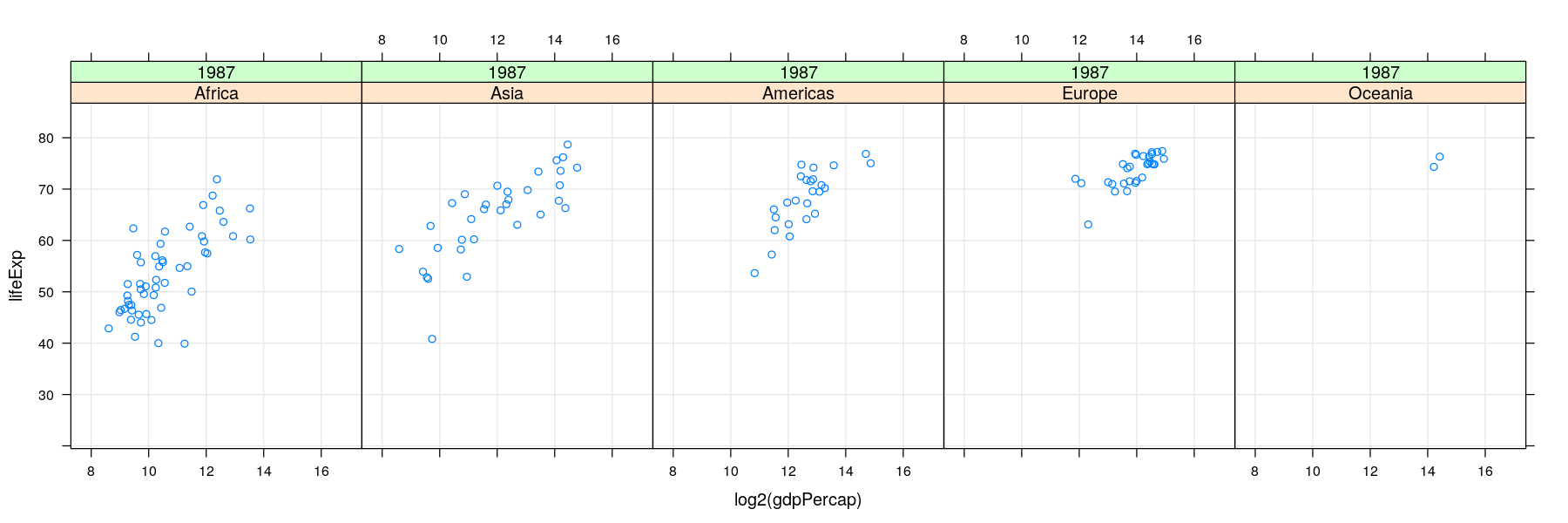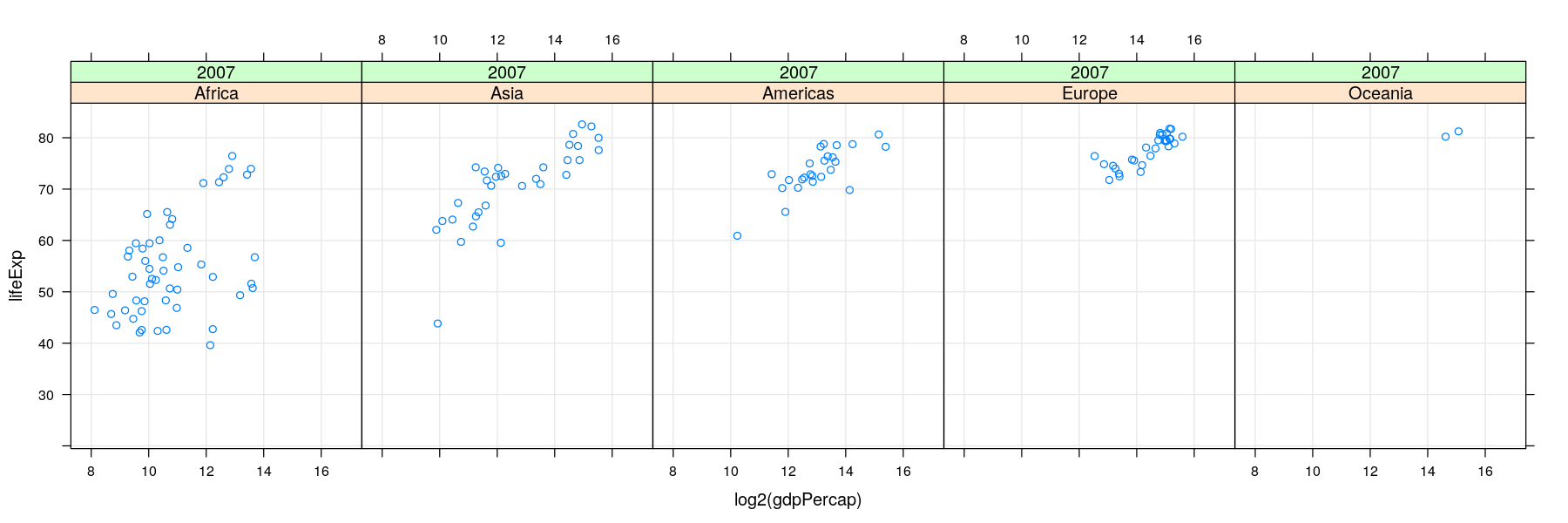
Time-series
- Essentially scatter plots with time on x-axis, conventionally with positions joined by lines
library(ggplot2)
vaccines <- transform(vaccines, count0 = ifelse(is.na(count), 0, count))
ggplot(data = vaccines, aes(x = year, y = sqrt(count0))) + geom_line(aes(group = state), alpha = 0.3)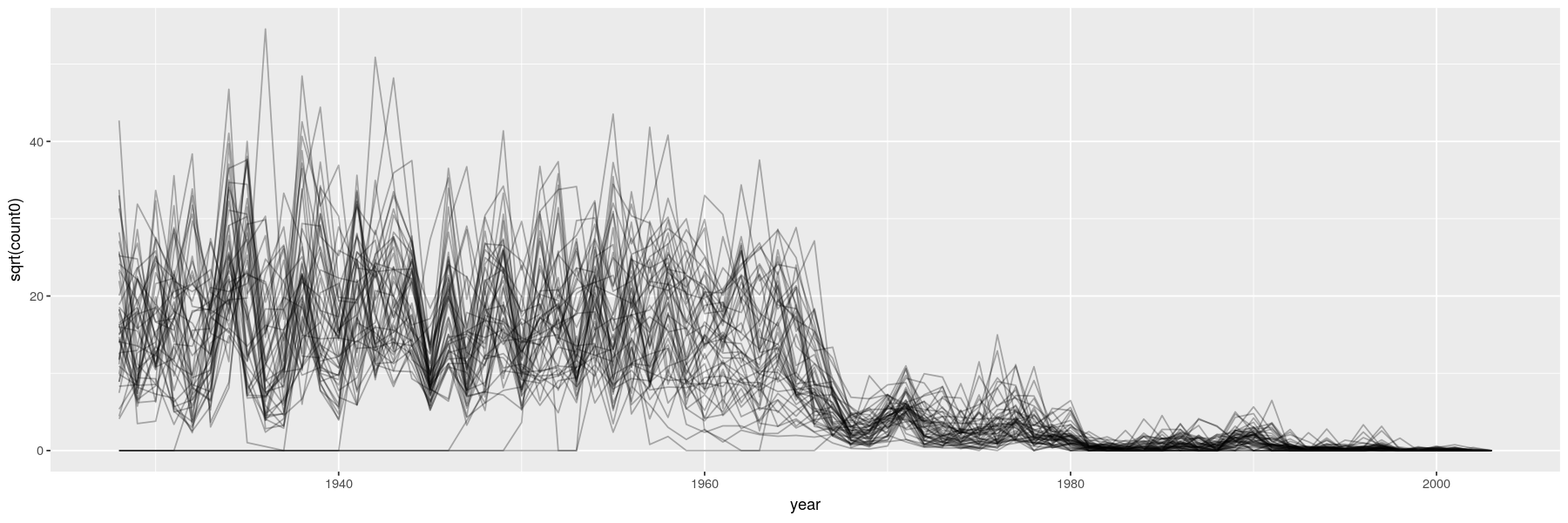
Time-series
- Essentially scatter plots with time on x-axis, conventionally with positions joined by lines
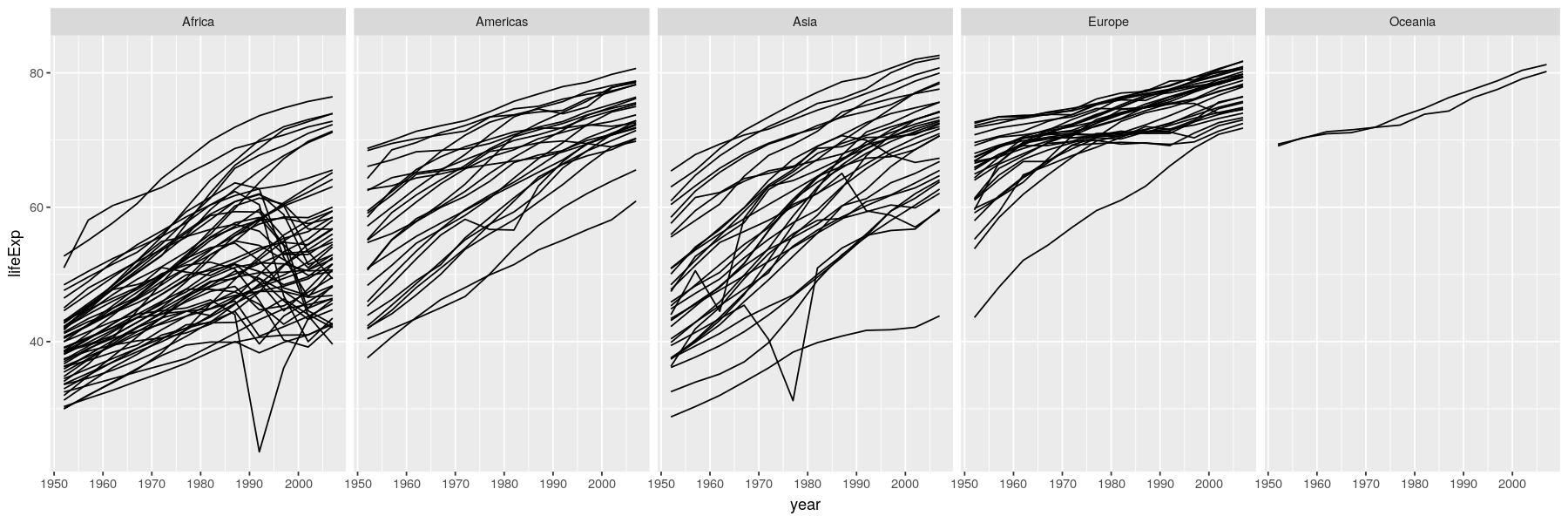
Dynamic and interactive graphs: past, present, and future
All these visualizations are static in nature
It would obviously be useful to have dynamic and interactive graphs
Traditionally, interactive graphs have had limited usefulness in reporting / publications
Cross-platform implementation is challenging
Implementations for interactive use available for a long time: e.g., xlisp-stat, Xgobi / Ggobi
Unfortunately, these did not gain mainstream popularity (difficult to install on Windows)
Dynamic and interactive graphs: past, present, and future
What has changed? Modern web technology: Javascript, always-available internet
Powerful and universally available on all platforms
Web-based reporting and publications becoming more acceptable
Challenges:
For security reasons, browsers limit access to local software
So not easy to connect browser with locally running software (like R)
Current state of interactive visualization using R
Many Javascript visualization libraries available
Several R packages provide interfaces to various libraries
These allow data manipulation in R, followed by visualization in a browser
More difficult to allow call-backs to R in response to user interaction
shinyfrom R Studio is the most powerful system currently available
Current state of interactive visualization using R
- I will end with two simple examples of interactions using the
plotlypackage
library(plotly)
p1 <- ggplot(data = gapminder, aes(x = year, y = lifeExp)) + geom_line(aes(group = country)) +
facet_grid(~ continent)
p2 <- ggplot(data = vaccines, aes(x = year, y = state, text = paste0("Measles: ", count, " per 100,100"))) +
geom_raster(aes(fill = sqrt(count0)))
w1 <- ggplotly(p1)
w2 <- ggplotly(p2)Printing
w1andw2will create the following plots in a browserplotlyis particularly attractive because it can easily convert mostggplot2plots