An Overview of the R Programming Environment
R for Clinical Trials
What is R?
An environment for statistical computing and graphics (from website)
Available as Free / Open Source Software
Very popular (both academia and industry)
Easy to try out on your own
Is it suitable for clinical trials research / analysis?
Yes! See this Task View if you are looking for specific methods not covered in the workshop
For regulatory issues, see the R certification page
Ask me if you still have any concerns after this workshop
Is R better than SAS?
I don’t know SAS, so I don’t have an opinion
Both are tools, you should use what you are comfortable with
Main differences: (from looking at some SAS examples for this workshop)
R is designed to be used interactively
It is more similar to traditional programming languages like C / Java / Python
Almost everything in R is done by calling functions (similar to SAS PROCs?)
Writing new functions is much easier (and very common) in R
Overall agenda for the workshop
Informal overview of R
More formal introduction to the language
Statistical analyis (model fitting, visualization, tests)
Case studies using clinical trial examples
Outline of first part: Overview
Installing R
Basics of using R
Example: Linear regresion
Working with “reproducible documents”
Installing R
R is most commonly used as a REPL (Read-Eval-Print-Loop)
When it is started, R Waits for user input
Evaluates and prints result
Waits for more input
There are several different interfaces to do this
R itself works on many platforms (Windows, Mac, UNIX, Linux)
Some interfaces are platform-specific, some work on most
R and the interface may need to be installed separately
I assume you have already done this! Otherwise:
Go to https://cran.r-project.org/ (or choose a mirror first)
Follow instructions depending on your platform (probably Windows)
This will install R, as well as a default graphical interface on Windows and Mac
I recommend a different interface called R Studio that needs to be installed separately
I hope that you have also done this (but it is not essential)
Running R
- Once installed, you can start the appropriate interface (or R directly) to get something like this:
R Under development (unstable) (2019-12-29 r77627) -- "Unsuffered Consequences"
Copyright (C) 2019 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
Natural language support but running in an English locale
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
Loading required package: utils
>
The
>represents a prompt indicating that R is waiting for input.The difficult part is to learn what to do next
The R REPL essentially works like a calculator
[1] 782[1] 3.857143[1] 7.389056[1] 1024
Formally, R evaluates the expression typed in at the prompt
This may sometimes result in an error (or a
+prompt requesting more input)
R has standard mathematical functions
[1] 25[1] 4.787492[1] 3628800[1] 15.10441
Most non-trivial tasks involve calling functions
[1] 3003[1] 3003[1] 1124250[1] NaNR supports variables
[1] 1024[1] 100[1] 3628800[1] 6.559763
Variable assignment is done using “ a <- b ” , but “ a = b ” also works
R can compute on vectors
[1] 15 [1] 0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 [1] 1 15 105 455 1365 3003 5005 6435 6435 5005 3003 1365 455 105 15 1
This is one of the most important distinguishing features of R
R has built-in functions for probability calculations
[1] 1.336346e-02 6.681731e-02 1.559070e-01 2.251991e-01 2.251991e-01 1.651460e-01 9.174777e-02 3.932047e-02
[9] 1.310682e-02 3.398065e-03 6.796131e-04 1.029717e-04 1.144130e-05 8.800998e-07 4.190952e-08 9.313226e-10 [1] 1.336346e-02 6.681731e-02 1.559070e-01 2.251991e-01 2.251991e-01 1.651460e-01 9.174777e-02 3.932047e-02
[9] 1.310682e-02 3.398065e-03 6.796131e-04 1.029717e-04 1.144130e-05 8.800998e-07 4.190952e-08 9.313226e-10
The results of any evaluation is usually printed, unless assigned to a variable
When printing vectors, R prefixes each output line with the index of the first element
R has functions that work on vectors
[1] 3.75[1] 3.75R can draw graphs
plot(x, p.x, ylab = "Probability", pch = 16)
title(main = sprintf("Binomial(%g, %g)", N, p))
abline(h = 0, col = "grey")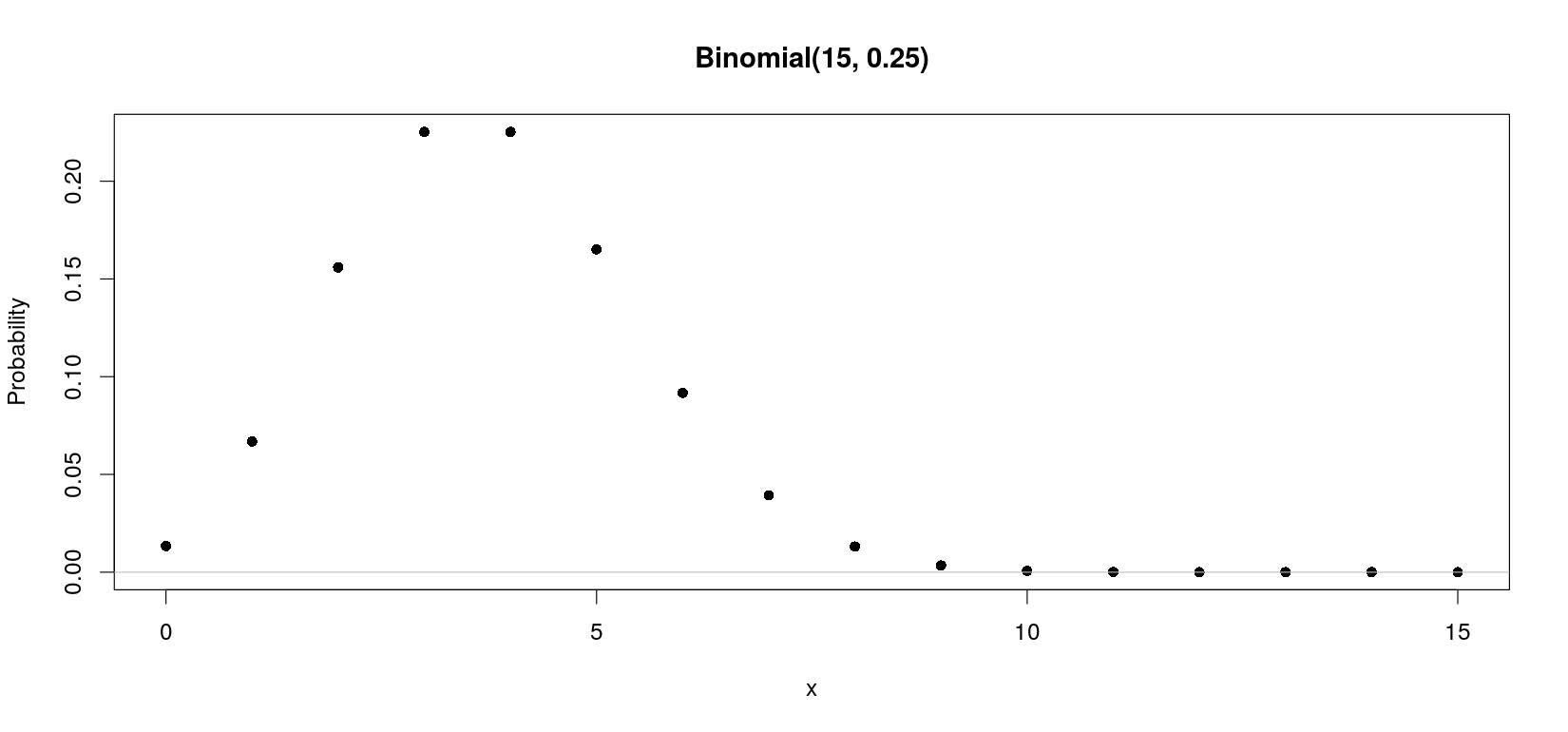
R can simulate random variables
[1] "H 1" "D 1" "C 1" "S 1" "H 2" "D 2" "C 2" "S 2" "H 3" "D 3" "C 3" "S 3" "H 4" "D 4" "C 4"
[16] "S 4" "H 5" "D 5" "C 5" "S 5" "H 6" "D 6" "C 6" "S 6" "H 7" "D 7" "C 7" "S 7" "H 8" "D 8"
[31] "C 8" "S 8" "H 9" "D 9" "C 9" "S 9" "H 10" "D 10" "C 10" "S 10" "H 11" "D 11" "C 11" "S 11" "H 12"
[46] "D 12" "C 12" "S 12" "H 13" "D 13" "C 13" "S 13" [1] "H 2" "H 6" "S 2" "C 12" "D 11" "H 4" "D 1" "C 9" "C 4" "H 5" "C 11" "D 3" "C 7" [1] "H 9" "H 1" "H 2" "C 8" "C 12" "S 6" "S 5" "S 4" "S 12" "C 10" "H 8" "C 9" "D 1" [1] 0.3719275 0.1936631 0.9836552 -0.4919062 -1.7152266 -0.9536651 0.9155696 1.5895887 -0.3702185
[10] 0.9767307 -1.1981943 0.6387483 -0.8821841 -1.3193350 -0.2383257 0.1816948 0.7416892 0.3211718
[19] -0.8479021 -0.4725004 0.6577830 1.4740185 1.3978639 -0.9053331 -1.3183413 -0.4472865 0.9149328
[28] 0.6456813 -0.2279315 0.4666840 0.5562906 1.9223093 -2.4408218 -1.0598998 -1.5190523 -0.4563874
[37] 0.2832823 -1.2485320 0.5840814 -2.1556053 -0.0695510 0.8110716 -1.9551105 -0.8455179 1.0413871
[46] -1.4115850 1.3930437 -0.9093232 -0.2552080 1.2633080[1] -0.1077754[1] 1.077932[1] -0.1487413R is in fact a full programming language
Variables
Functions
Control flow structures
For loops, while loops
If-then-else (branching)
Distinguishing features
Focus on vectors and vectorized operations
Treatment of functions at par with other object types
We will see a few examples to illustrate what I mean by this
Example: Linear regression
Let us simulate some fake height-weight data
ht <- rnorm(200, mean = 172, sd = 10) # height in cm
bmi <- rnorm(200, mean = 22, sd = 2.2) # bmi (independent of height)
wt <- bmi * (ht / 100)^2 # weight in kgA simple least squares regression model is fit using the lm() function
As usual, no output is printed is the result is assigned to a variable
Examine fitted model
Useful output is produced by the functions coef() and summary()
(Intercept) ht
-70.1440360 0.7871475
Call:
lm(formula = wt ~ ht)
Residuals:
Min 1Q Median 3Q Max
-26.0490 -4.8284 0.1839 4.5992 17.3154
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -70.14404 8.33442 -8.416 7.73e-15 ***
ht 0.78715 0.04845 16.246 < 2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 6.744 on 198 degrees of freedom
Multiple R-squared: 0.5714, Adjusted R-squared: 0.5692
F-statistic: 263.9 on 1 and 198 DF, p-value: < 2.2e-16You should always plot the data to assess model fit!
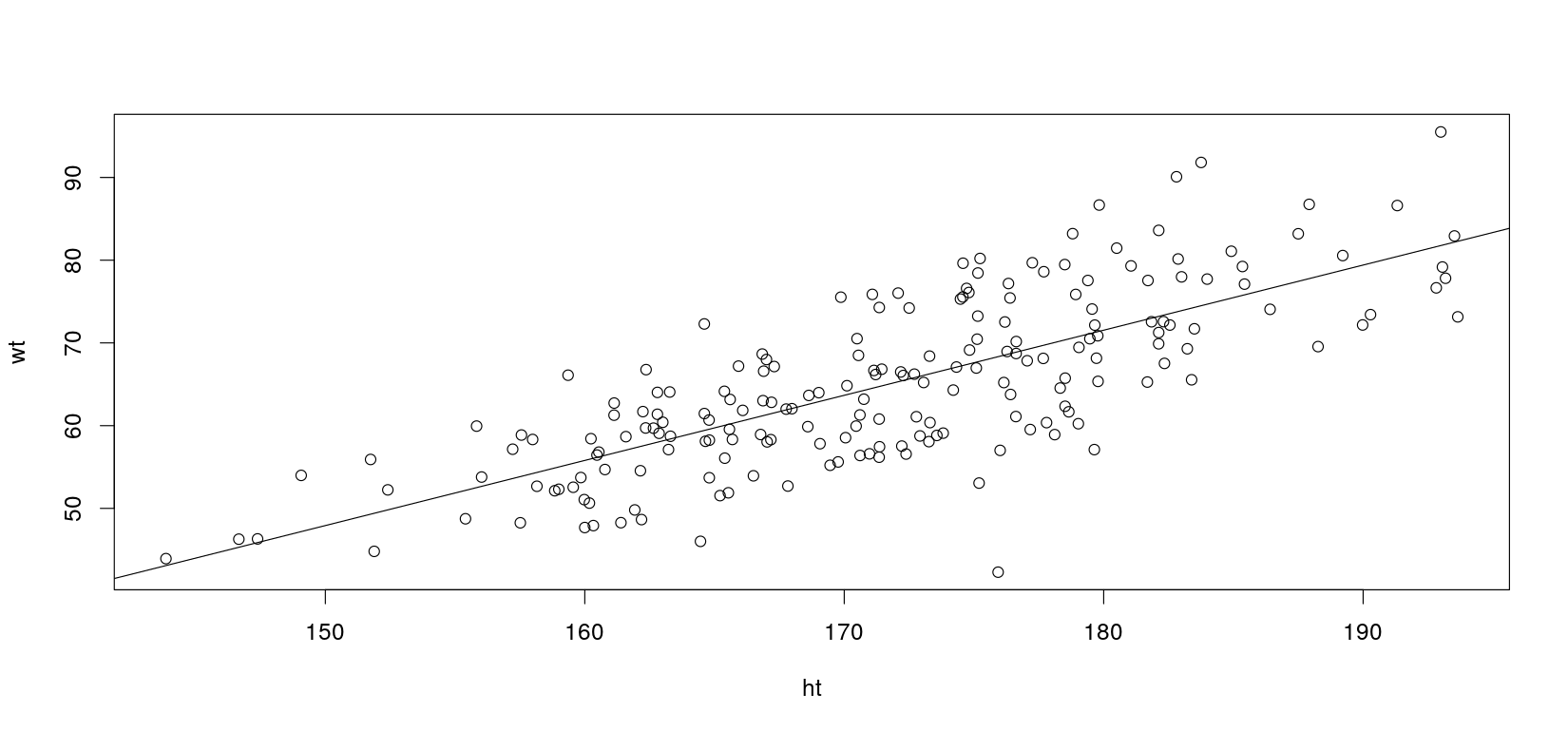
Let’s introduce a mistake in the data
Switch height and weight for the first case
Fit the OLS regression line again
Call:
lm(formula = wt ~ ht)
Residuals:
Min 1Q Median 3Q Max
-23.914 -6.989 -1.586 5.451 121.831
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 42.07680 12.61916 3.334 0.00102 **
ht 0.13714 0.07352 1.865 0.06361 .
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 12.73 on 198 degrees of freedom
Multiple R-squared: 0.01727, Adjusted R-squared: 0.01231
F-statistic: 3.48 on 1 and 198 DF, p-value: 0.06361Plot the data and regression line again
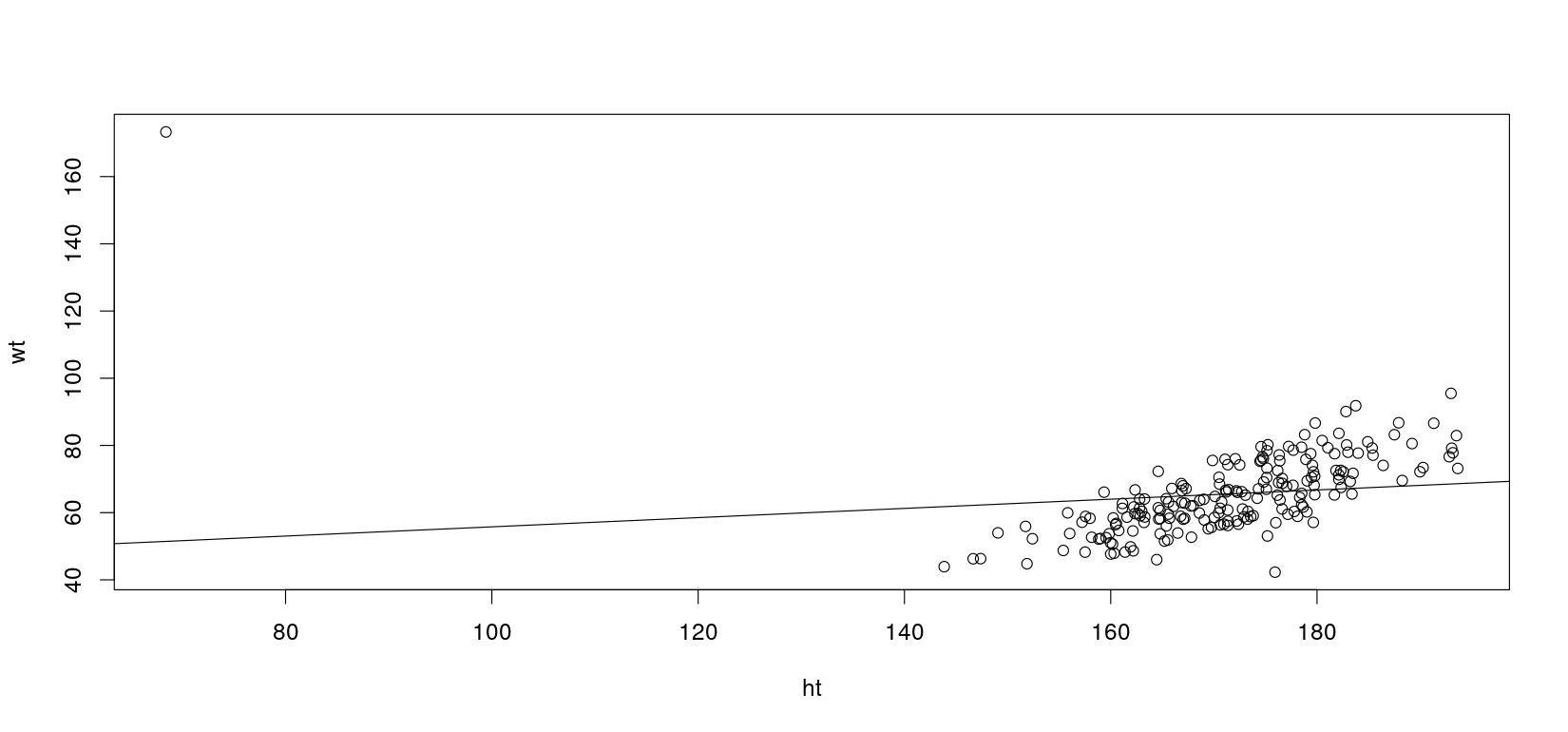
How can we “fix” this?
Two common approaches:
Delete “outlier” and re-fit model
Use robust regression
Least squares fit is sensitive to outliers
Instead, minimise sum of absolute errors (or something similar)
How?
We can find and use a method someone has already implemented
We can implement a solution of our own
R package ecosystem
R gives access to an extensive toolset
Most standard data analysis methods are already implemented
Can be extended by writing add-on packages
Thousands of add-on packages are available from CRAN
Quality varies
This may matter in regulated environments
For this workshop, I will mostly use the “default” R packages
These come by default along with a standard installation of R
Consist of “base” packages
[1] "base" "compiler" "datasets" "graphics" "grDevices" "grid" "methods" "parallel"
[9] "splines" "stats" "stats4" "tcltk" "tools" "utils"
- … and “recommended” packages
[1] "boot" "class" "cluster" "codetools" "foreign" "KernSmooth" "lattice" "MASS"
[9] "Matrix" "mgcv" "nlme" "nnet" "rpart" "spatial" "survival"
There are many other extremely powerful packages that you can explore later
The other option: implement our own solution
This is often surprisingly easy
We will come back to the robust regression example later
Interactive vs “pipeline” analysis
R encourages an interactive data analysis workflow
Result of each step should dictate the next step to take
Understanding this will help make sense of the design of R
Sometimes it is useful to apply a standard workflow on a new dataset
These are usually done using “R scripts”
I would also strongly encourage using dynamic “notebooks”
Notebooks in R Studio
Notebooks allow you to mix text and code
Notebook only contains code, results are dynamically generated
R Studio supports several variants
These slides are written using “R Markdown”
Open the file names 01-roverview.rmd in R Studio
These can also be used to generate HTML / PDF / Word reports (requires some more software)
Benefit: reproducible results, no copy-paste errors
Before we move on…
Please spend some time getting comfortable with R Studio:
Load the R Markdown file in R Studio
Run the “code chunks” (marked using
```{r} ... ```)Useful shortcut: Ctrl + Shift + Enter
Try making small changes and re-run