Lattice Graphics: Custom Panel Functions and Layers
Deepayan Sarkar
Use case: displaying model fits
Common use case: we often want to augment a raw data plot with a model fit
We have already seen one basic example: add a regression line
Depending on data, we may be interested in something more complicated
We will discuss some possible approaches
We restrict our attention to regression models (continuous response)
We assume familiarity with model-fitting in R (including
summary(),fitted(),predict(), etc.)
Datasets for illustration
Indometh: Pharmacokinetics of indometacin on six subjects over eleven time points.
Subject time conc
1 1 0.25 1.50
2 1 0.50 0.94
3 1 0.75 0.78
4 1 1.00 0.48
5 1 1.25 0.37
6 1 2.00 0.19
7 1 3.00 0.12
8 1 4.00 0.11
9 1 5.00 0.08
10 1 6.00 0.07
11 1 8.00 0.05
12 2 0.25 2.03
13 2 0.50 1.63
14 2 0.75 0.71
15 2 1.00 0.70
16 2 1.25 0.64
17 2 2.00 0.36
18 2 3.00 0.32
19 2 4.00 0.20
20 2 5.00 0.25
21 2 6.00 0.12
22 2 8.00 0.08Datasets for illustration
xyplot(conc ~ time | Subject, data = Indometh, strip = FALSE, layout = c(6, 1),
pch = 16, grid = TRUE, xlab = "Time since indometacin injection (hr)",
ylab = "Plasma concentrations of \n indometacin (mcg/ml)")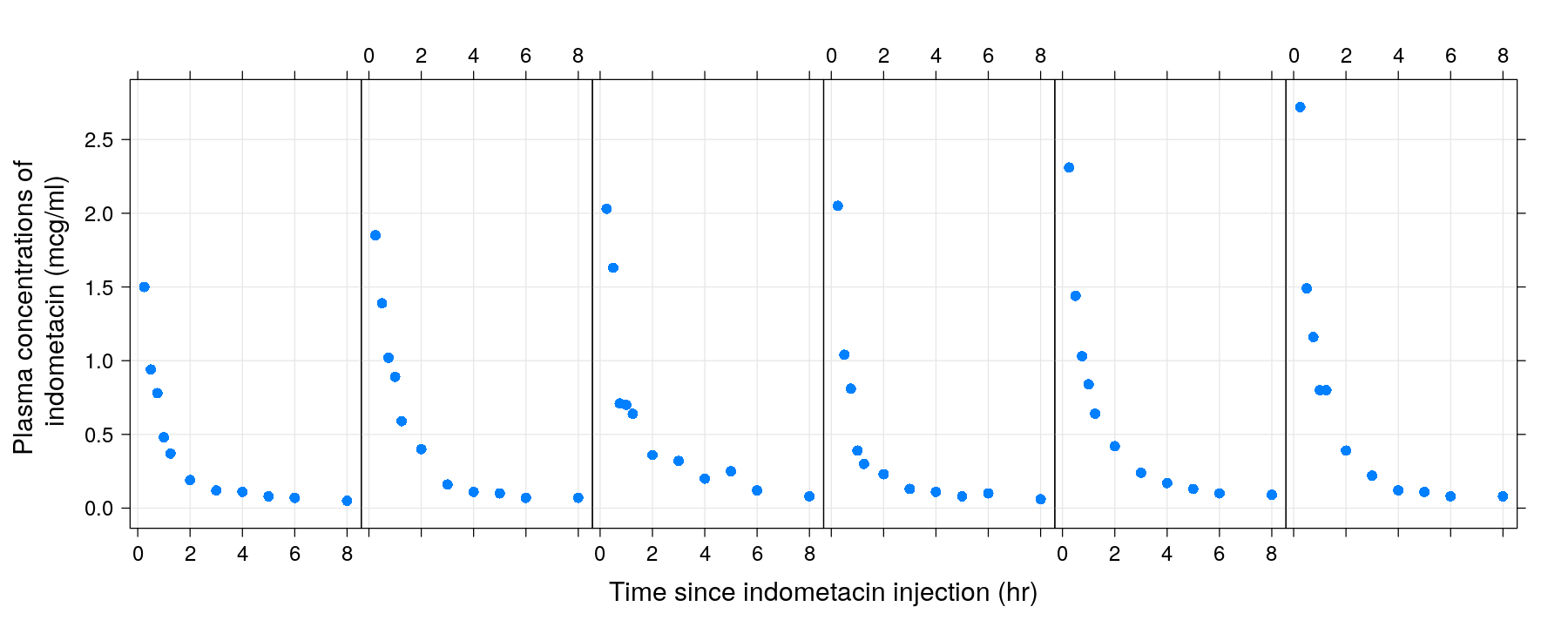
Datasets for illustration
Gcsemvfrom themlmRevpackage.Records the GCSE exam scores in a science subject.
Marks in two components (coursework and written paper) are recorded separately.
Interested in modeling the expected written score based on course work score, taking into account the gender of the student.
school student gender written course
1 20920 16 M 23 NA
2 20920 25 F NA 71.2
3 20920 27 F 39 76.8
4 20920 31 F 36 87.9
5 20920 42 M 16 44.4
6 20920 62 F 36 NADatasets for illustration
xyplot(written ~ course | gender, data = Gcsemv, grid = TRUE,
xlab = "Coursework score", ylab = "Written exam score")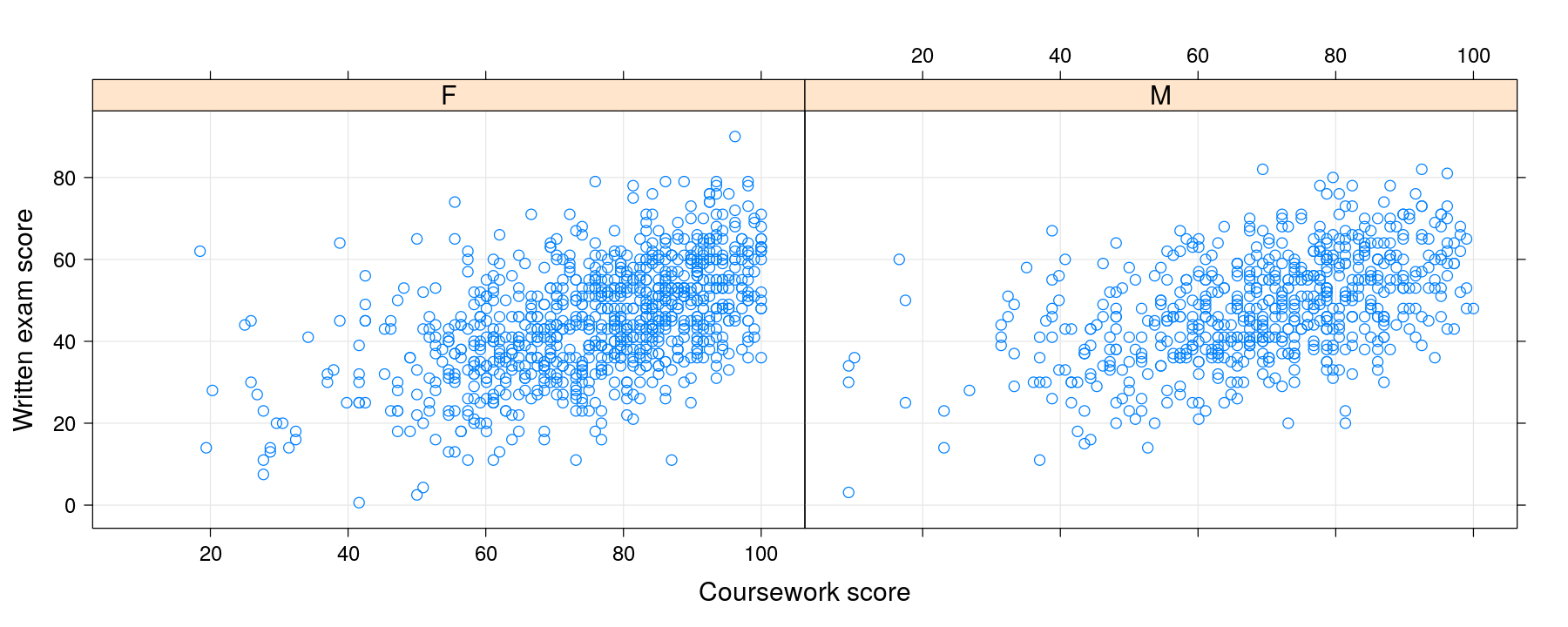
Datasets for illustration
- This dataset has many missing observations
school student gender written course
68137 : 104 77 : 14 F:1128 Min. : 0.60 Min. : 9.25
68411 : 84 83 : 14 M: 777 1st Qu.:37.00 1st Qu.: 62.90
68107 : 79 53 : 13 Median :46.00 Median : 75.90
68809 : 73 66 : 13 Mean :46.37 Mean : 73.39
22520 : 65 27 : 12 3rd Qu.:55.00 3rd Qu.: 86.10
60457 : 54 110 : 12 Max. :90.00 Max. :100.00
(Other):1446 (Other):1827 NA's :202 NA's :180
- These are not shown in the previous plot
Adding smoothers
The default panel function
panel.xyplot()supports adding two basic smoothers:linear regression
loess
This is done using the
type=argument, which is in fact more generally useful, e.g.,type = "p": pointstype = "l": linestype = "b": bothtype = "h": ‘histogram-like’ linestype = "r": linear regression linetype = "a": line joining per-group average (categorical predictor)type = "smooth": loess line
Multiple values of
type=can be combined, e.g.,type = c("p", "smooth")
Adding smoothers
xyplot(written ~ course | gender, data = Gcsemv, grid = TRUE,
type = c("p", "smooth"), col.line = "black")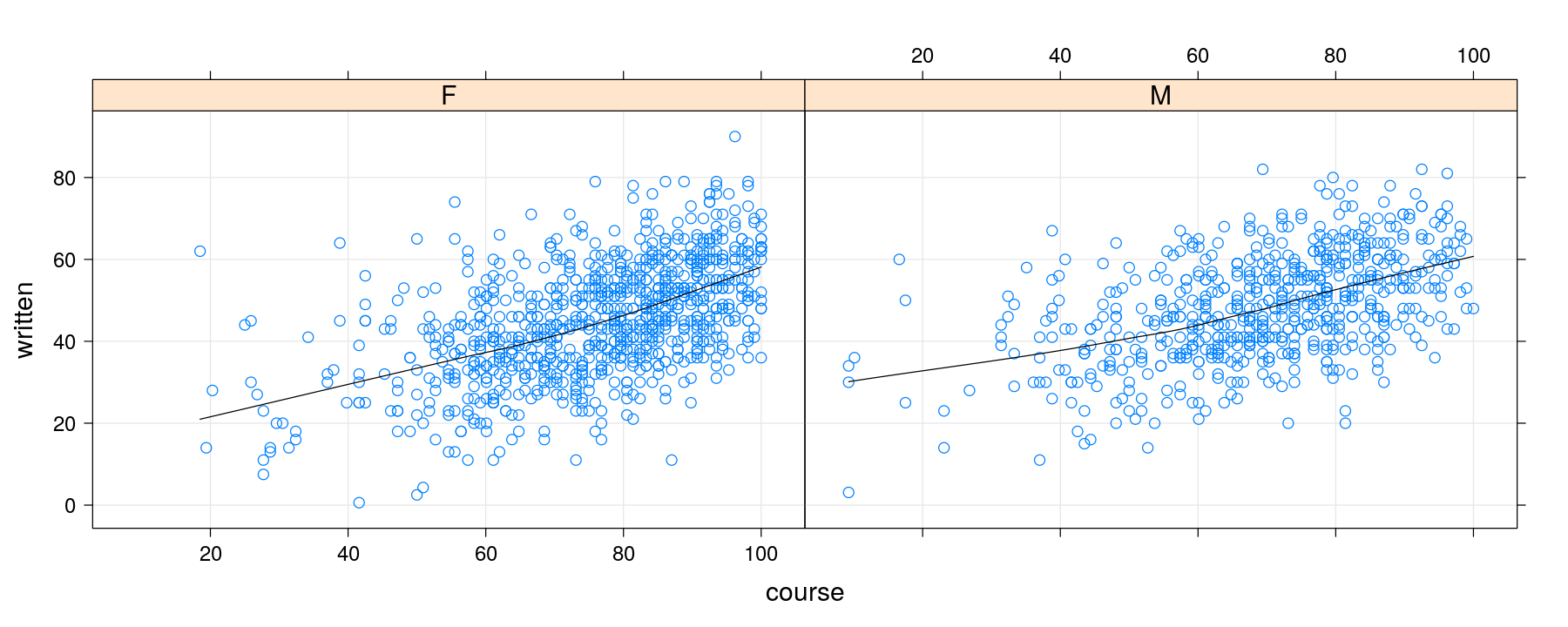
Adding smoothers
To display other kinds of model fits, we need a different panel function function
We can either use an alternative available elsewhere, or write our own
Traditional R graphics plots are customized by adding components incrementally
This is done using “low-level” functions such as
lines(),points(),text(), etc.The analogue in lattice is to write panel functions.
Panel functions
Panel functions are just like other R functions:
accepts some input arguments
does something
terminates with a return value
Responsible for actually drawing the graphical content
Every lattice plot has a panel function
Gets executed every time a panel is drawn, with input data specific to that panel
As the input data is different for each panel, so is the result
Many built-in panel functions are already available in lattice and latticeExtra
The most important of these are the default panel functions
Custom panel functions are usually created by combining two or more of them
Understanding how panel functions work
Understanding how panel functions work
Every high level function has a default panel function
xyplot()has default panel functionpanel.xyplot()This default can be replaced by a user-speficied alternative using the
panel=argumentThe default panel function is of course a valid (though uninteresting) choice
So the following call is equivalent to the earlier one
- The following call is also equivalent (ask if it is not clear why!)
Understanding how panel functions work
To do anything interesting, we need access to the data
These are supplied as argument to the panel function
The names of the arguments depends on the specific panel function
For
panel.xyplot(), the data arguments arexandySo another equivalent call is
Understanding how panel functions work
We are now ready to add some non-trivial components; for example,
A reference grid before the data is plotted using
panel.grid()A loess fit using
panel.loess()Marginal “rugs” to indicate missing components using
panel.rug()
xyplot(written ~ course | gender, data = Gcsemv,
panel = function(x, y, ...) {
panel.grid(h = -1, v = -1)
panel.xyplot(x, y, ...)
panel.loess(x, y, ..., col = "black")
panel.rug(x = x[is.na(y)], y = y[is.na(x)])
})- For repeated use, it may be helpful to name the function instead of defining it inline
Understanding how panel functions work
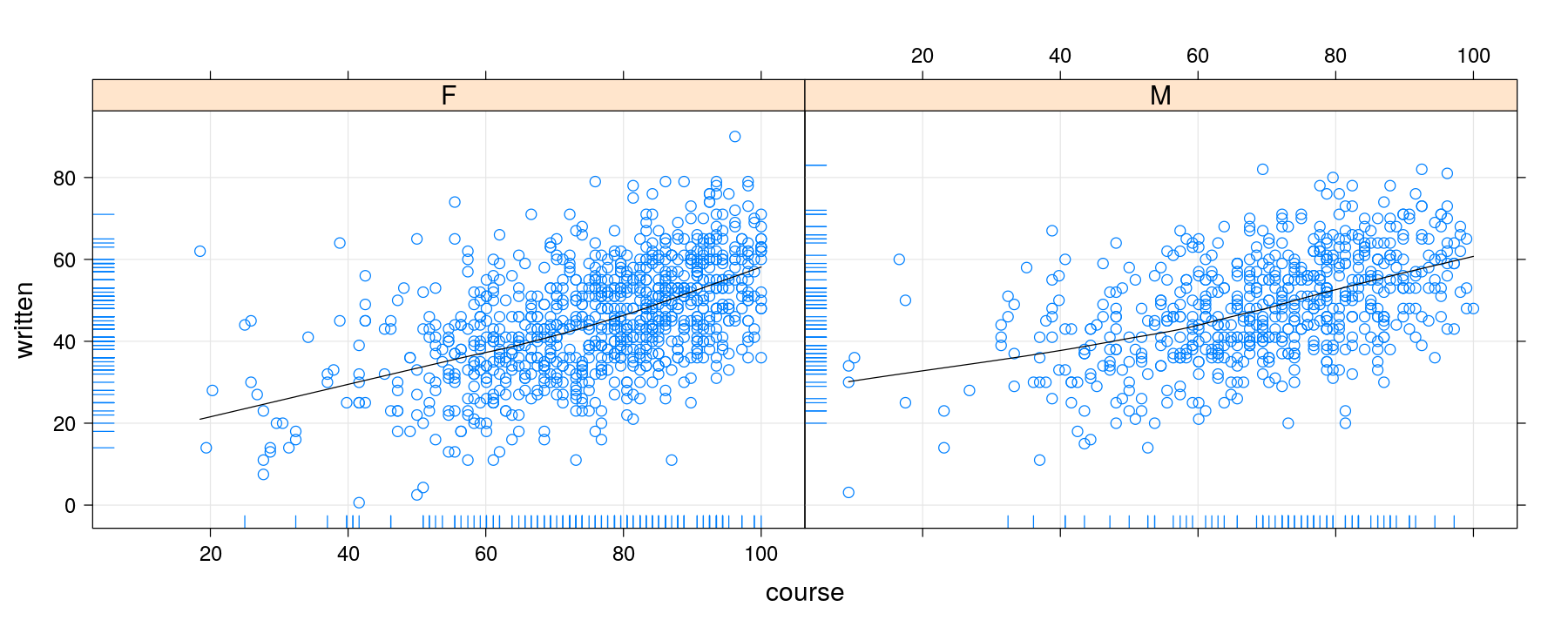
Understanding how panel functions work
Individual elements are added by predefined component functions
panel.grid(): background gridpanel.xyplot(): actual data pointspanel.loess(): the loess smoothpanel.rug(): the marginal rugs indicating missing values
Together they define a procedure for plotting the data in a panel
Ordering of components determines order of plotting
The ... argument
One important feature here is the use of
...R functions normally have (0, 1, or more) named arguments
Can also optionally have a special
...argumentSuch functions can be supplied extra arguments not matching the named arguments
These extra arguments are usually passed on to further function calls
In fact, this is why optional arguments of panel functions can be supplied in high-level calls
The ... argument
- As noted earlier,
panel.xyplot()itself directly supports these enhancements (except the rug)
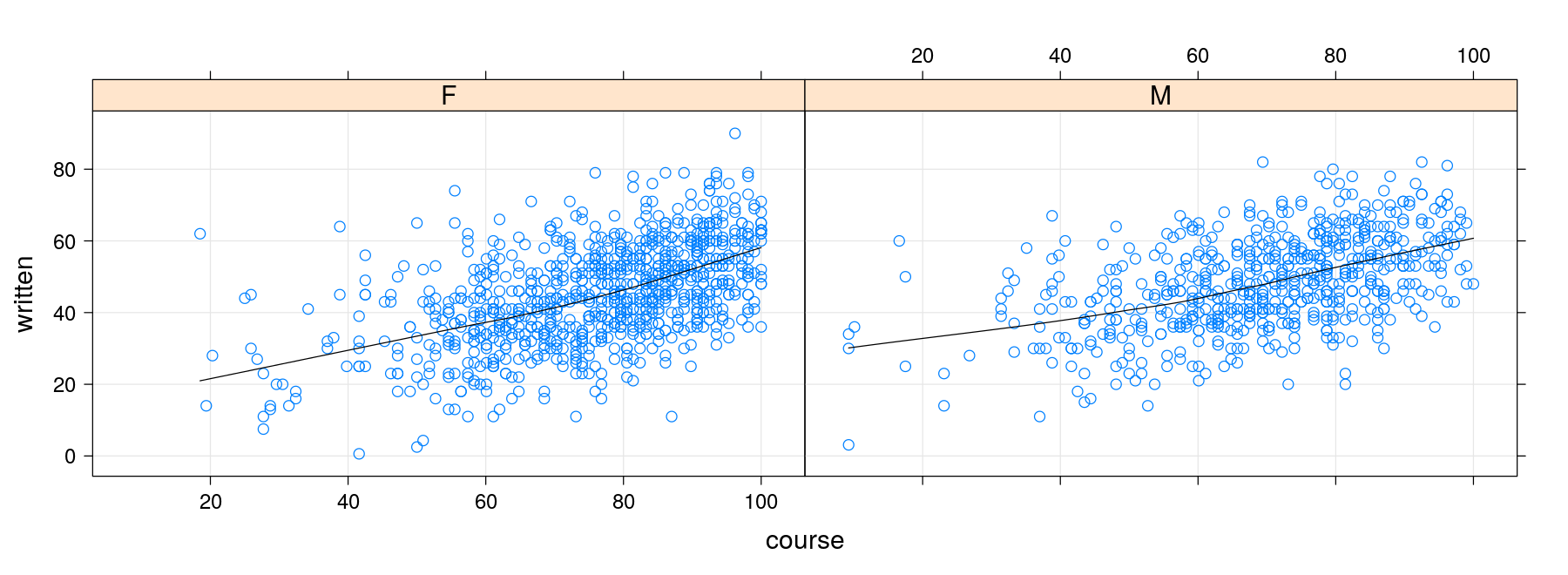
xyplot(written ~ course | gender, data = Gcsemv,
panel = function(x, y) {
panel.xyplot(x, y, grid = TRUE, type = c("p", "smooth"), col.line = "black")
})
The ... argument
xyplotuses the...mechanism to pass arguments to its panel functionSo these arguments can be directly supplied to
xyplot()
xyplot(written ~ course | gender, data = Gcsemv, grid = TRUE,
type = c("p", "smooth"), col.line = "black")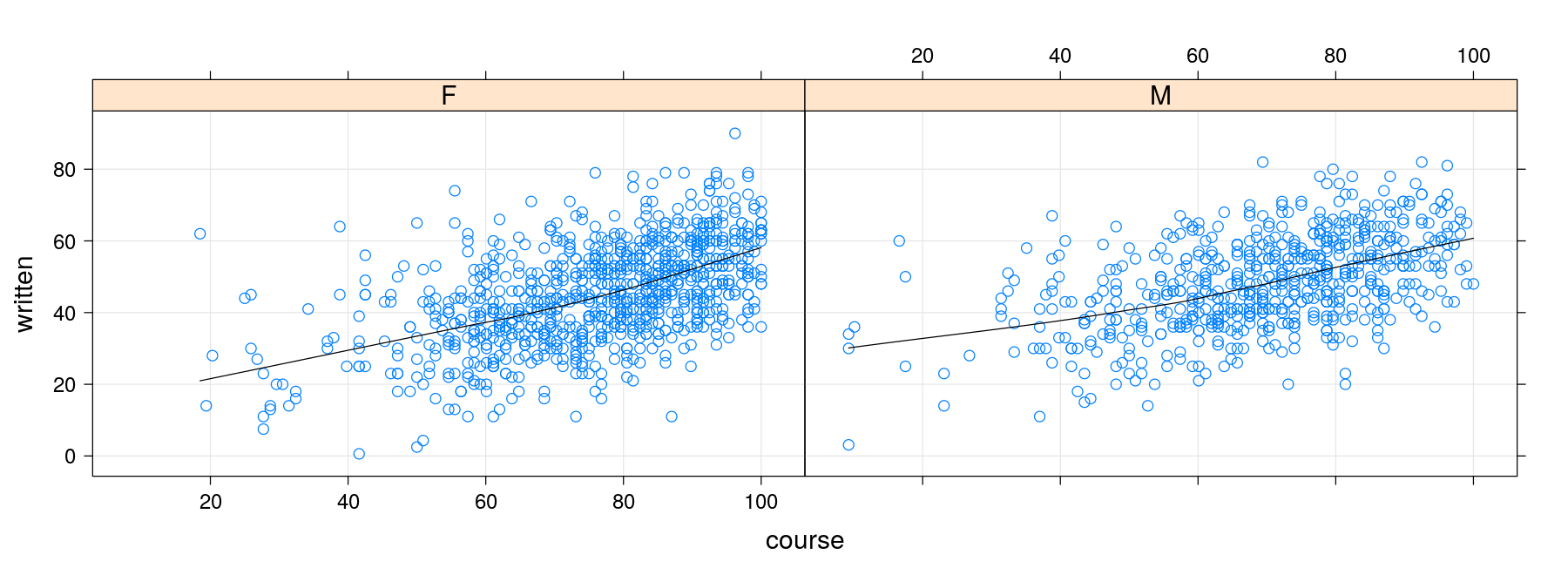
Alternative approach using layers
Thanks to Felix Andrews, latticeExtra provides an alternative layering mechanism
Inspired by ggplot2
This basically works by adding layers to the default panel display
layer()adds a call after the default panel functionlayer_()adds a call before the default panel functionglayer()andglayer_()add group-specific layers
Each such call contains an expression that could appear in a panel function
Alternative approach using layers
library(latticeExtra)
p <- xyplot(written ~ course | gender, data = Gcsemv, grid = TRUE)
p + layer(panel.loess(x, y, col = "black")) + layer(panel.rug(x[is.na(y)], y[is.na(x)]))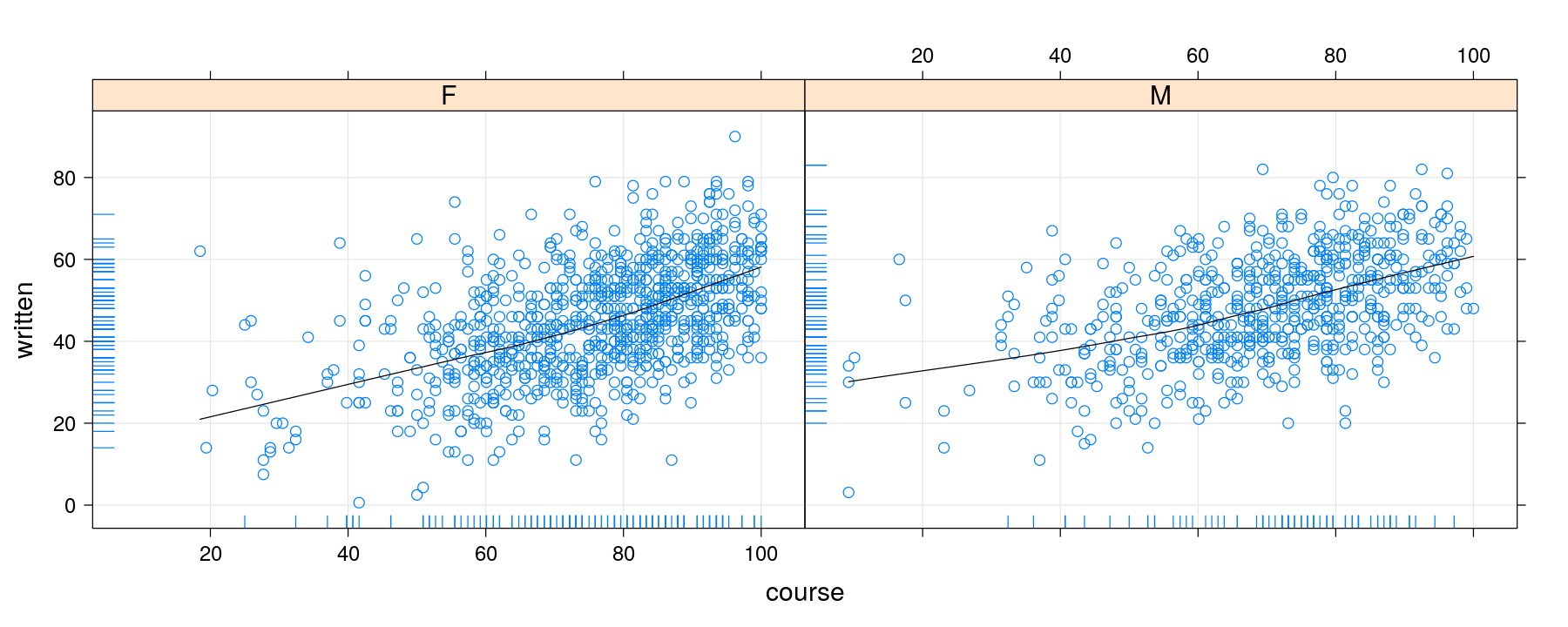
Alternative approach using layers
p <- xyplot(written ~ course | gender, data = Gcsemv, grid = TRUE)
p + layer(panel.smoother(x, y, method = "loess")) + layer(panel.rug(x[is.na(y)], y[is.na(x)]))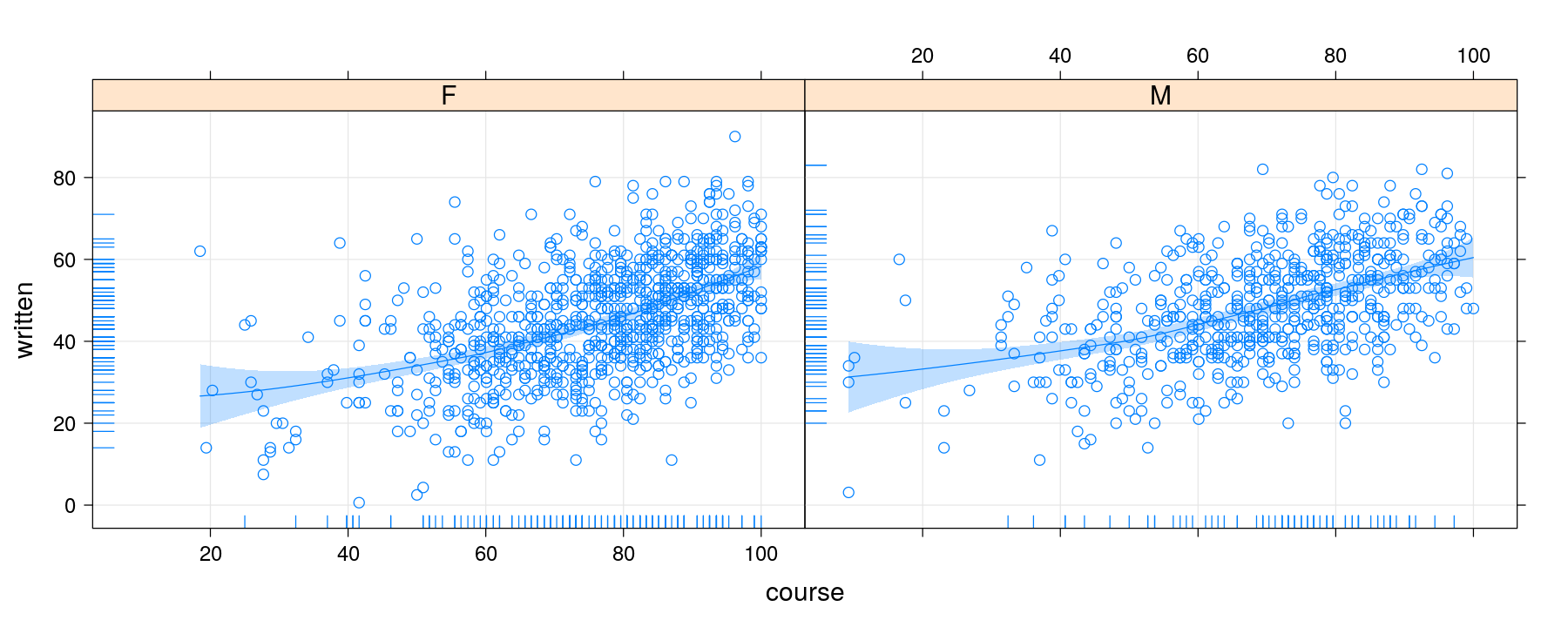
Alternative approach using layers
p <- xyplot(written ~ course | gender, data = Gcsemv, grid = TRUE)
p + layer(panel.smoother(x, y, method = "lm", form = y ~ poly(x, 2)))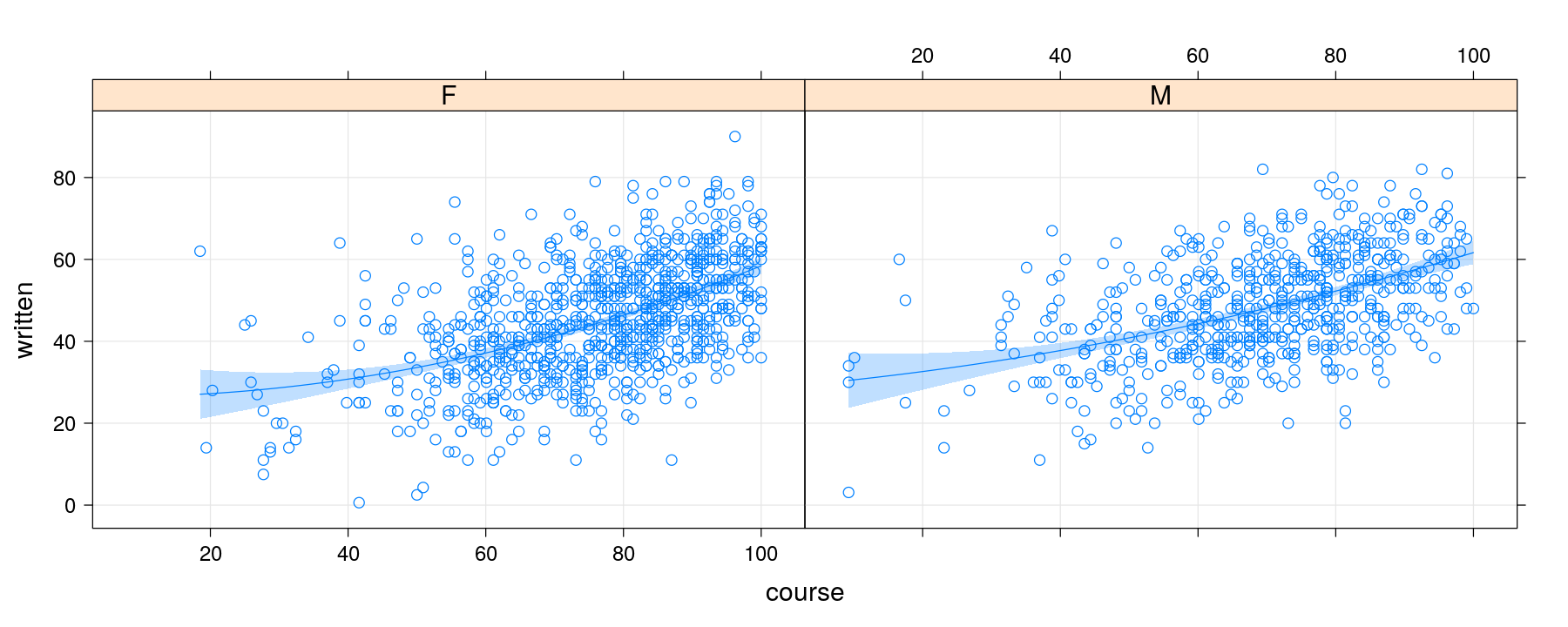
Alternative approach using layers
panel.smoother()is a powerful interface for a wide range of modelsWorks for any formula-based modeling functions
Assumes existence of
predict()method with standard interface
The layer mechanism is very convenient for simple customizations
Works only when suitable component panel functions are already available
Makes use of non-standard evaluation, so should be used with care
These examples also emphasize one feature we have not discussed earlier
High-level lattice functions create an object (rather than the final plots)
These objects have (S3) class “trellis”
The actual plotting is done by the corresponding
print()methodThe object can be manipulated in several ways before plotting
Fitting non-standard models
p <- xyplot(conc ~ time | Subject, data = Indometh, strip = FALSE,
layout = c(6, 1), pch = 16, grid = TRUE)
p + layer(panel.abline(lm(y ~ x), col = "grey40")) # clearly inappropriate model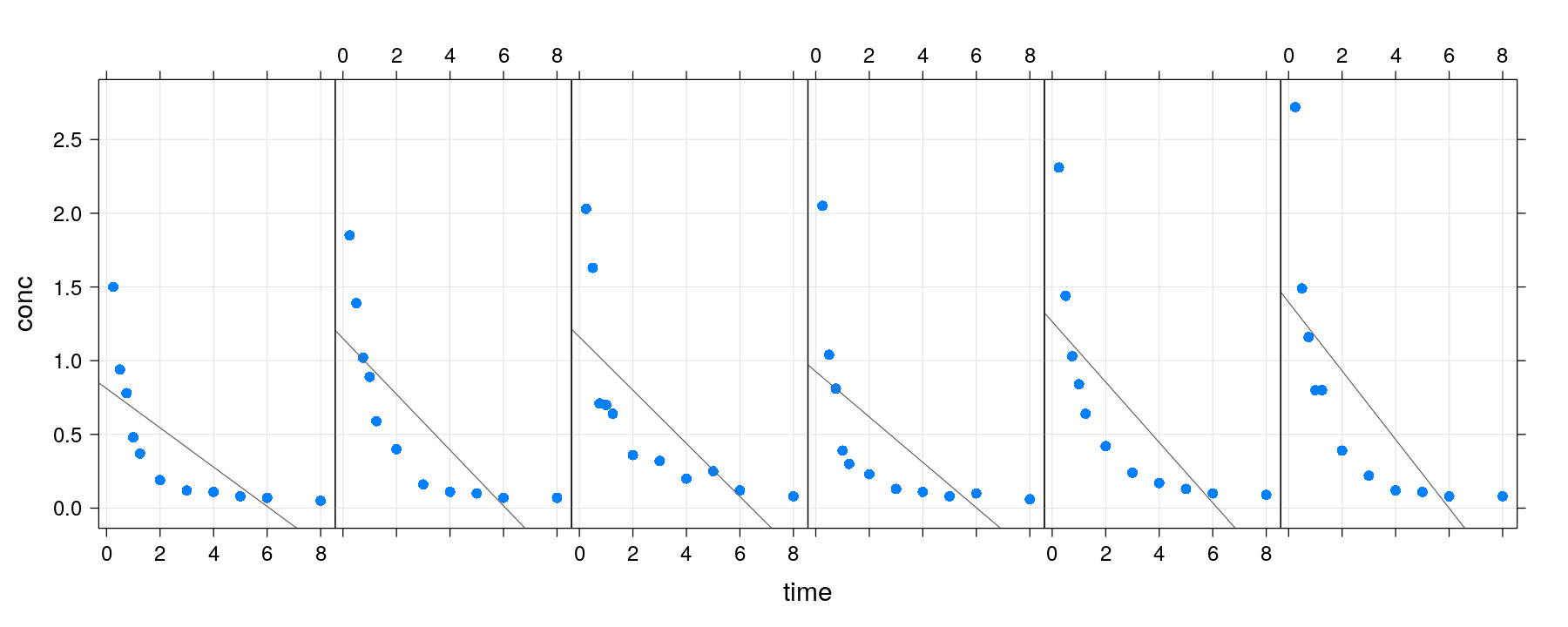
Fitting non-standard models
- Without thinking too much, try a simple linear regression model with
1/timeas predictor.
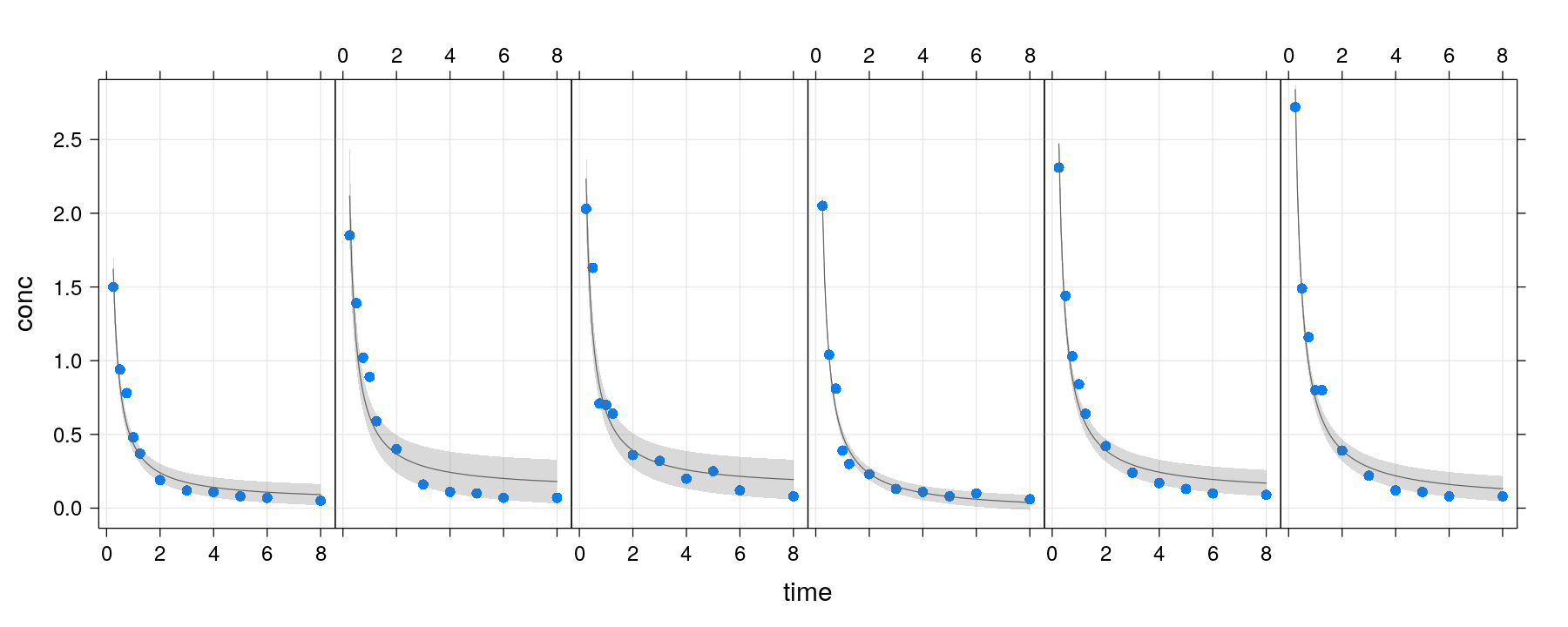
Fitting non-standard models
The specific model used here is not important
We can replace the call to
lm()with any other formula-based modeling functionsFor example, this data is used to illustrate the nonlinear biexponential model (see
help(SSbiexp))
\[ E(y_t) = \alpha_1 \exp(-\phi_1 t) + \alpha_2 \exp(-\phi_2 t)\,,\,\phi_1 > \phi_2 > 0. \]
Fitting non-standard models
p + layer(panel.smoother(x, y, method = "nls",
form = y ~ SSbiexp(x, A1, lrc1, A2, lrc2), se = FALSE))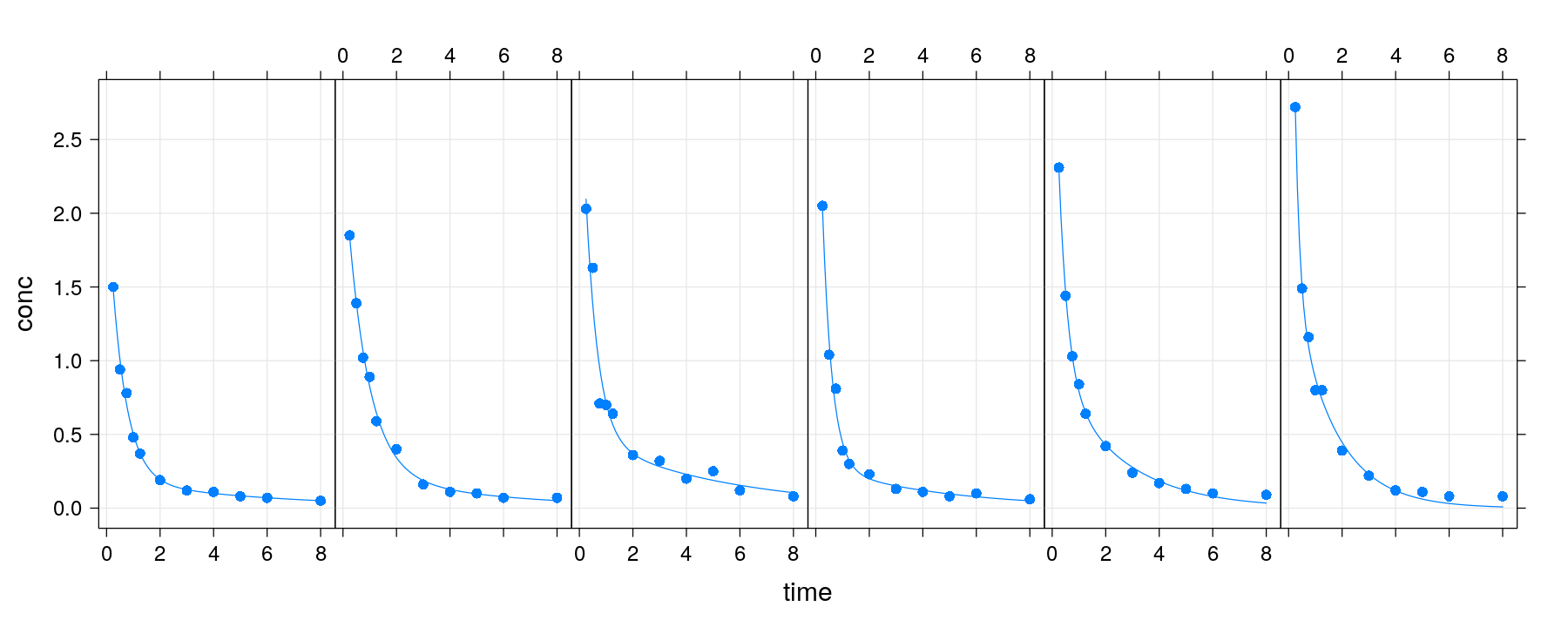
Limitations of this approach
Only works for panel-specific models
We are often interested in more complex models that use the full data
To use such models, one usually fits the model separately, before attempting to create the plot
- Example: A nonlinear mixed effects model generalizing the earlier per-subject model
\[ E(y_{it}) = (\alpha_1 + a_{1i}) \exp(-(\phi_1 + p_{1i}) t) + (\alpha_2 + a_{2i}) \exp(-(\phi_2 + p_{2i}) t) \]
Here
\(i\) indexes subjects,
\(t\) denotes time of measurement,
random effects \(a_{1i}, p_{1i}, a_{2i}, p_{2i}\) assumed to be independent,
parameters in the model are \(\alpha_1, \phi_1, \alpha_2, \phi_2\), and the variances of the corresponding random effects.
Per-subject fixed effect model
Call:
Model: conc ~ SSbiexp(time, A1, lrc1, A2, lrc2) | Subject
Data: Indometh
Coefficients:
A1 lrc1 A2 lrc2
1 2.029277 0.5793887 0.1915474 -1.7877849
4 2.198132 0.2423123 0.2545223 -1.6026860
2 2.827673 0.8013195 0.4989175 -1.6353512
5 3.566103 1.0407660 0.2914970 -1.5068522
6 3.002250 1.0882119 0.9685230 -0.8731358
3 5.468312 1.7497936 1.6757521 -0.4122004
Degrees of freedom: 66 total; 42 residual
Residual standard error: 0.0755502 A1 lrc1 A2 lrc2
1 2.029277 0.5793887 0.1915474 -1.7877849
4 2.198132 0.2423123 0.2545223 -1.6026860
2 2.827673 0.8013195 0.4989175 -1.6353512
5 3.566103 1.0407660 0.2914970 -1.5068522
6 3.002250 1.0882119 0.9685230 -0.8731358
3 5.468312 1.7497936 1.6757521 -0.4122004Nonlinear mixed effects model
(fm.mixed <- nlme(conc ~ SSbiexp(time, A1, lrc1, A2, lrc2), data = Indometh,
random = pdDiag(A1 + lrc1 + A2 + lrc2 ~ 1)))Nonlinear mixed-effects model fit by maximum likelihood
Model: conc ~ SSbiexp(time, A1, lrc1, A2, lrc2)
Data: Indometh
Log-likelihood: 54.59671
Fixed: list(A1 ~ 1, lrc1 ~ 1, A2 ~ 1, lrc2 ~ 1)
A1 lrc1 A2 lrc2
2.8275372 0.7736221 0.4614716 -1.3441022
Random effects:
Formula: list(A1 ~ 1, lrc1 ~ 1, A2 ~ 1, lrc2 ~ 1)
Level: Subject
Structure: Diagonal
A1 lrc1 A2 lrc2 Residual
StdDev: 0.5714106 0.1580778 0.1115978 8.539108e-06 0.08149341
Number of Observations: 66
Number of Groups: 6 A1 lrc1 A2 lrc2
1 2.087866 0.8013747 0.3500886 -1.344102
4 2.261839 0.5425485 0.4733169 -1.344102
2 2.756906 0.8022321 0.5485764 -1.344102
5 3.240318 0.9715604 0.3295015 -1.344102
6 2.989983 0.7461872 0.5386318 -1.344102
3 3.628311 0.7778296 0.5287143 -1.344102Goal: Use fitted model to augment plot
Two approaches:
Make model available to panel function and calculate fitted model inside
Calculate fitted values before plotting anything
First approach: calulate fitted values inside panel function
panel.biexp <- function(x, y, fit, ...)
{
panel.xyplot(x, y, ...)
xx <- do.breaks(current.panel.limits()$xlim, 50)
submodel <- coef(fit)[packet.number(), , drop = FALSE]
yy <- with(submodel,
A1 * exp(-exp(lrc1) * xx) + A2 * exp(-exp(lrc2) * xx))
panel.lines(xx, yy, col = "black")
}current.panel.limits()gives current axis limitspacket.number()tells us whichSubjectis represented in panelUsed to extract corresponding coefficients as single-row data frame / list
Requires knowledge of model that has been fit (so not generally useful)
First approach: calulate fitted values inside panel function
xyplot(conc ~ time | Subject, data = Indometh, strip = FALSE, layout = c(6, 1), pch = 16,
grid = TRUE, fit = fm.fixed, panel = panel.biexp) # fixed model provided as 'fit'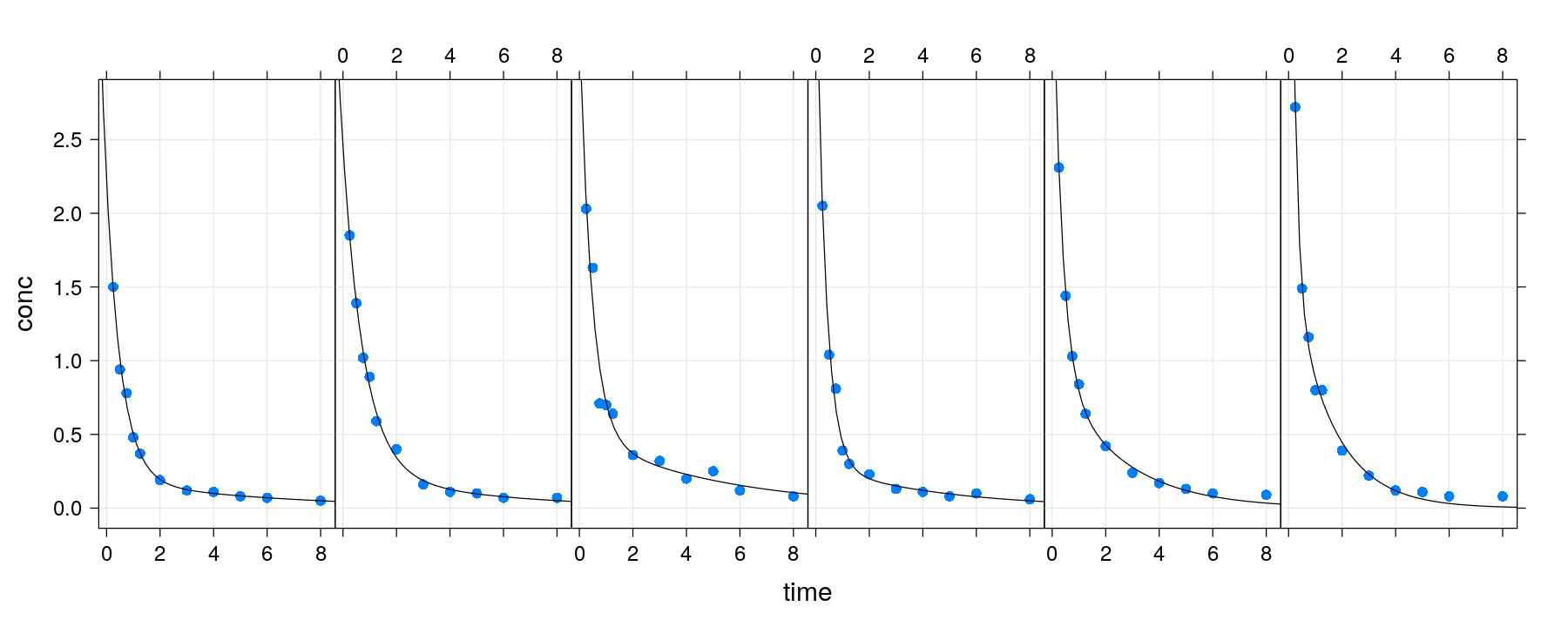
First approach: calulate fitted values inside panel function
xyplot(conc ~ time | Subject, data = Indometh, strip = FALSE, layout = c(6, 1), pch = 16,
grid = TRUE, fit = fm.mixed, panel = panel.biexp) # mixed model provided as 'fit'
Second approach: calculate fitted values beforehand
fitted()method extracts fitted values from model.Corresponds to the full dataset, not specific to panel.
panel.fitpred <- function(x, y, ..., fit1, fit2, subscripts)
{
panel.xyplot(x, y, ...)
panel.lines(x, fit1[subscripts], col = "grey60")
panel.lines(x, fit2[subscripts], col = "black")
}subscriptsindexes rows of the original dataset that end up in the panel
Second approach: calculate fitted values beforehand
- Fitted values provided as extra arguments (unfortunately no non-standard evaluation)
xyplot(conc ~ time | Subject, Indometh, strip = FALSE, layout = c(6, 1), pch = 16, grid = TRUE,
fit1 = Indometh$fitted.fixed, fit2 = Indometh$fitted.mixed, panel = panel.fitpred)
Second approach: calculate fitted values beforehand
- Again, the layering approach can simplify this kind of augmentation
pdata <- xyplot(conc ~ time | Subject, Indometh, strip = FALSE, layout = c(6, 1), pch = 16, grid = TRUE)
pfixed <- xyplot(fitted.fixed ~ time | Subject, data = Indometh, type = "l", col = "grey60")
pmixed <- xyplot(fitted.mixed ~ time | Subject, data = Indometh, type = "l", col = "black")
pdata + pfixed + pmixed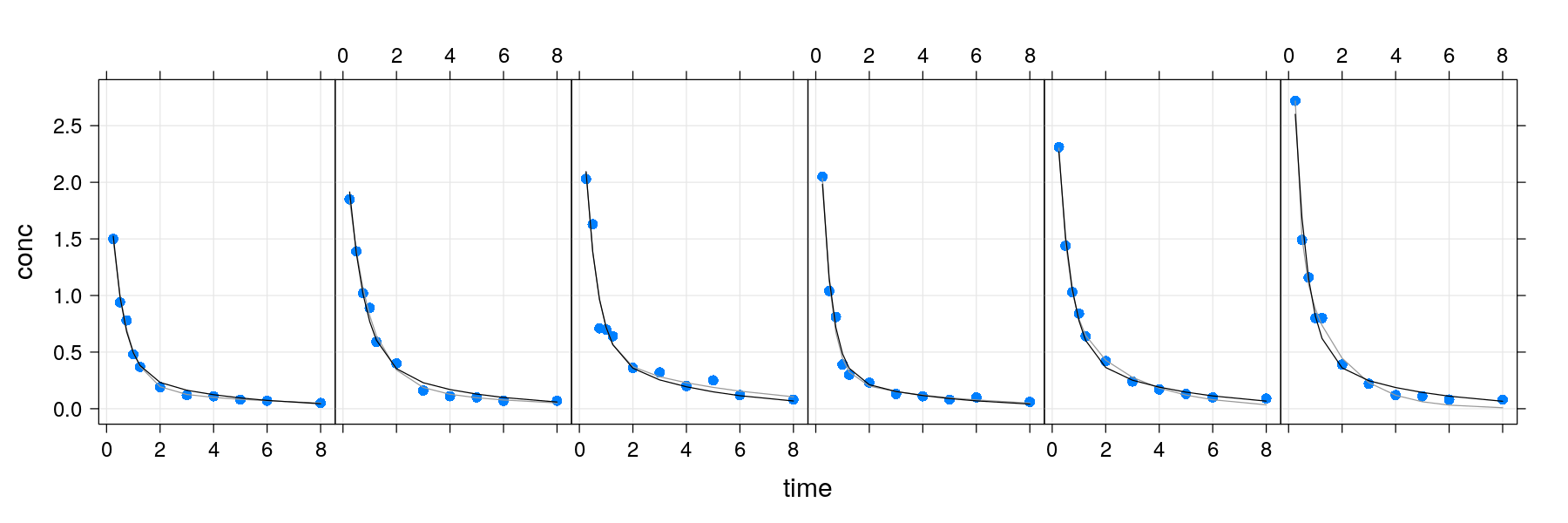
Second approach: calculate fitted values beforehand
In fact, the dataset need not be same in the plots being combined
Consider three models for the
Gcsemvdata set
fm.common <- lm(written ~ poly(course, 2), data = subset(Gcsemv, !(is.na(written) | is.na(course))))
fm.additive <- update(fm.common, written ~ poly(course, 2) + gender)- Next, construct a dataset to contain predicted values
Second approach: calculate fitted values beforehand
pdata <- xyplot(written ~ course | gender, data = Gcsemv, grid = TRUE)
pfitted <- xyplot(written.common + written.additive ~ course | gender, data = grid,
type = "l", col = c("black", "red"))
pdata + pfitted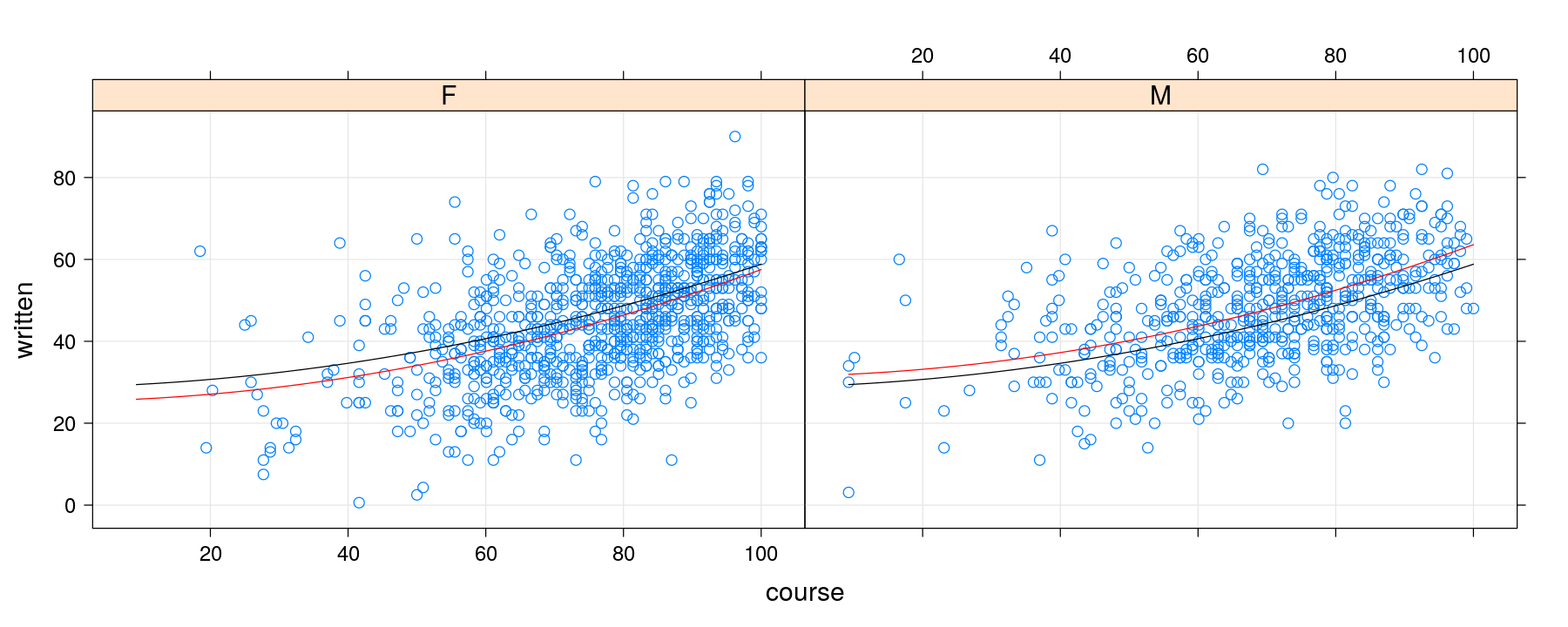
Second approach: calculate fitted values beforehand
update(pdata + pfitted, key = list(text = list(c("common", "additive")),
lines = list(col = c("black", "red")), columns = 2))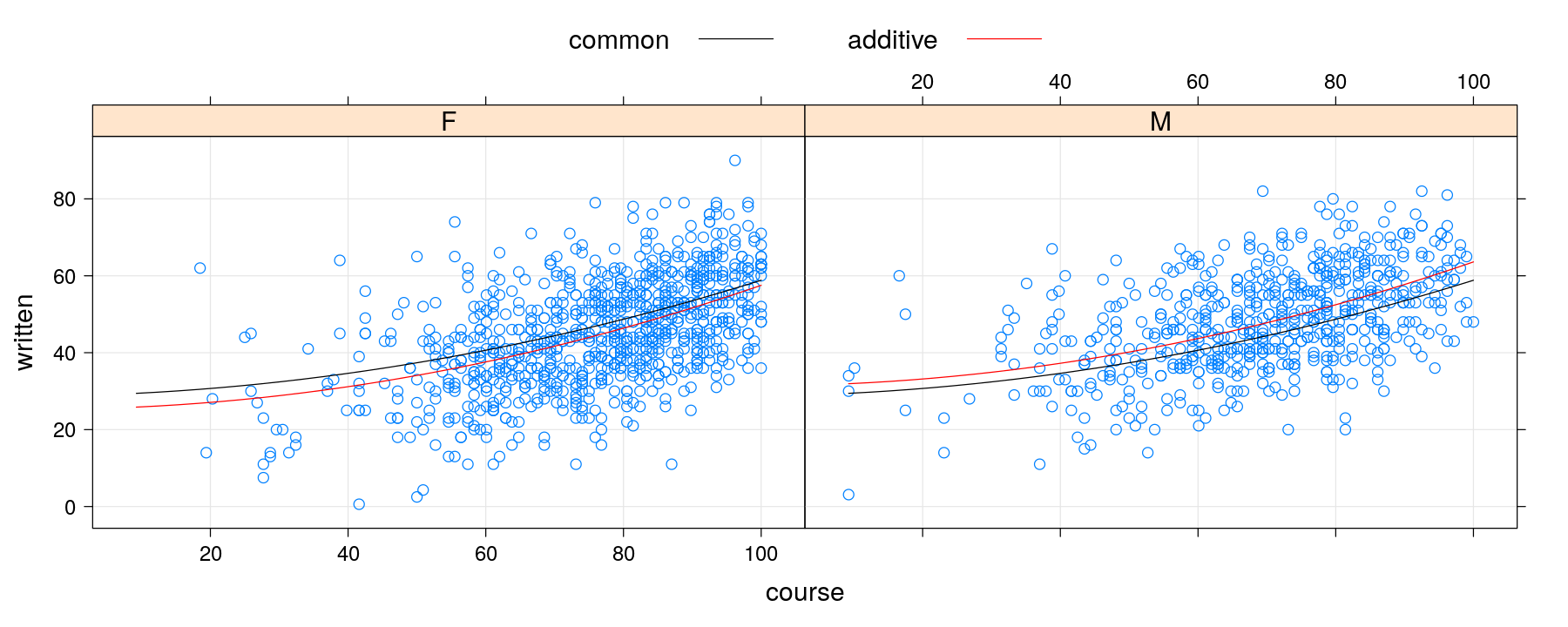
More examples of layering
data(Chem97, package = "mlmRev")
p <- histogram(~ gcsescore | factor(score) + gender, data = Chem97, type = "density",
border = NA, col = hcl.colors(1, "Pastel 1"))
p + layer(panel.curve(dnorm(x, mean = mean(x), sd = sd(x)))) ## fails (too much non-standard evaluation)
More examples of layering
panel.dnorm <- function(x, ...) {
m <- mean(x); s <- sd(x)
panel.curve(dnorm(x, mean = m, sd = s), ...)
}
p + layer(panel.dnorm(x, col = "grey40"))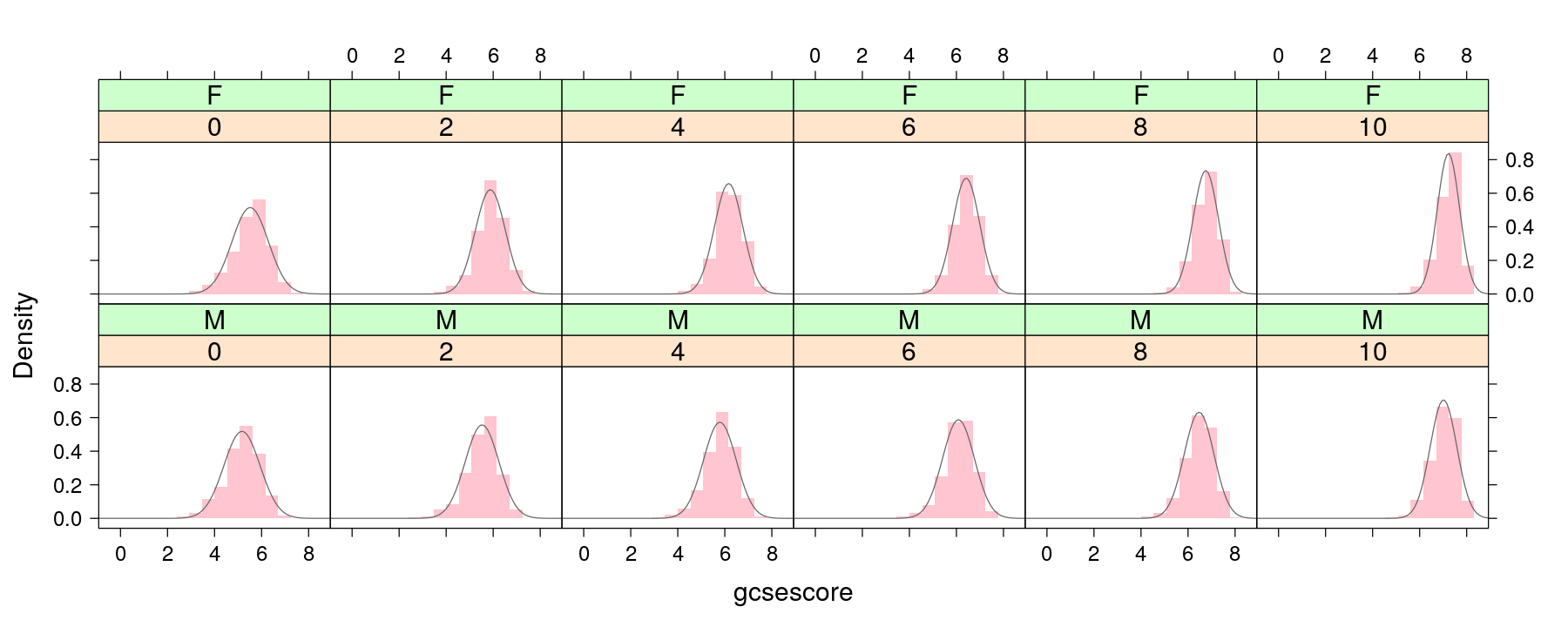
More examples of layering
VADeathsDF <- as.data.frame.table(VADeaths, responseName = "Rate") # 1940 Death rates in Virginia, USA
barchart(Rate ~ Var1 | Var2, VADeathsDF, origin = 0) +
layer(panel.text(x, y, labels = as.character(y), pos = 1, cex = 0.75))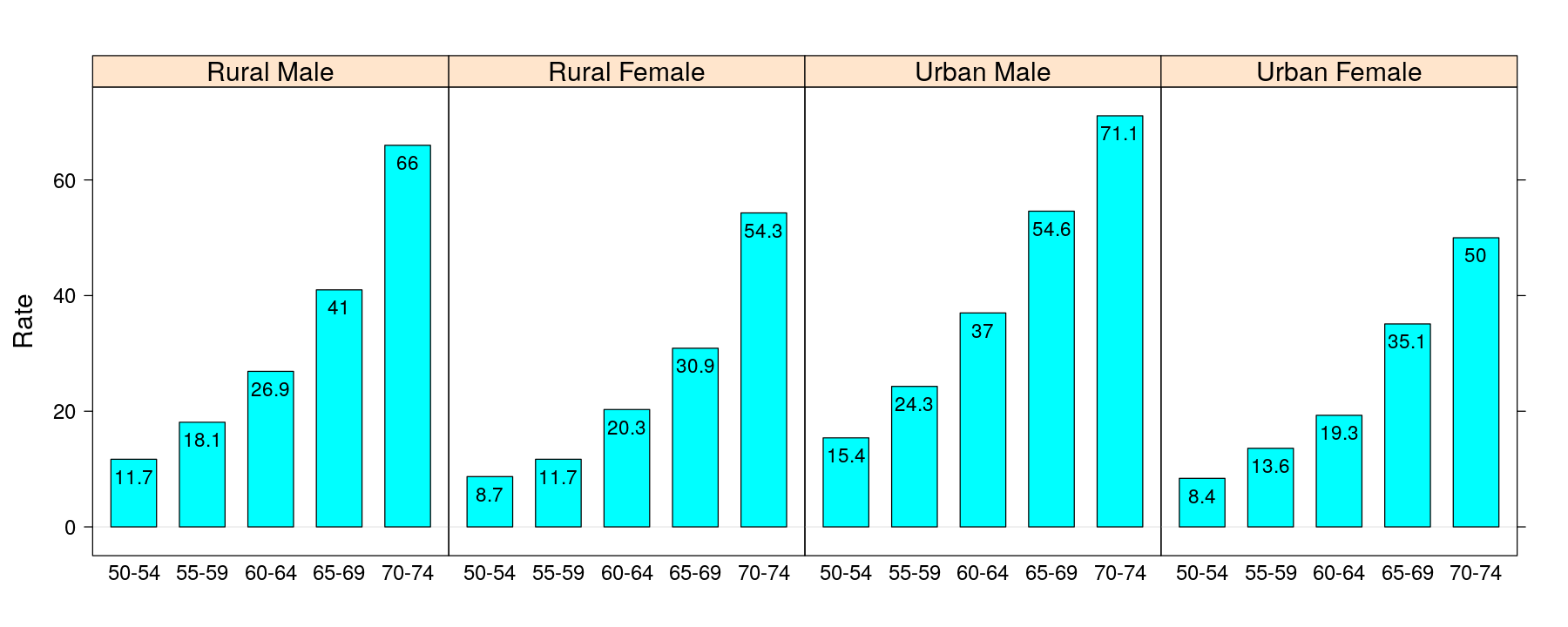
More examples of layering
dotplot(Var1 ~ Rate, VADeathsDF, groups = Var2, pch = 16, type = "o", aspect = 0.75) +
glayer(panel.text(x[5], y[5], group.value, srt = 30, cex = 0.7, adj = 0.75, col = "grey50"))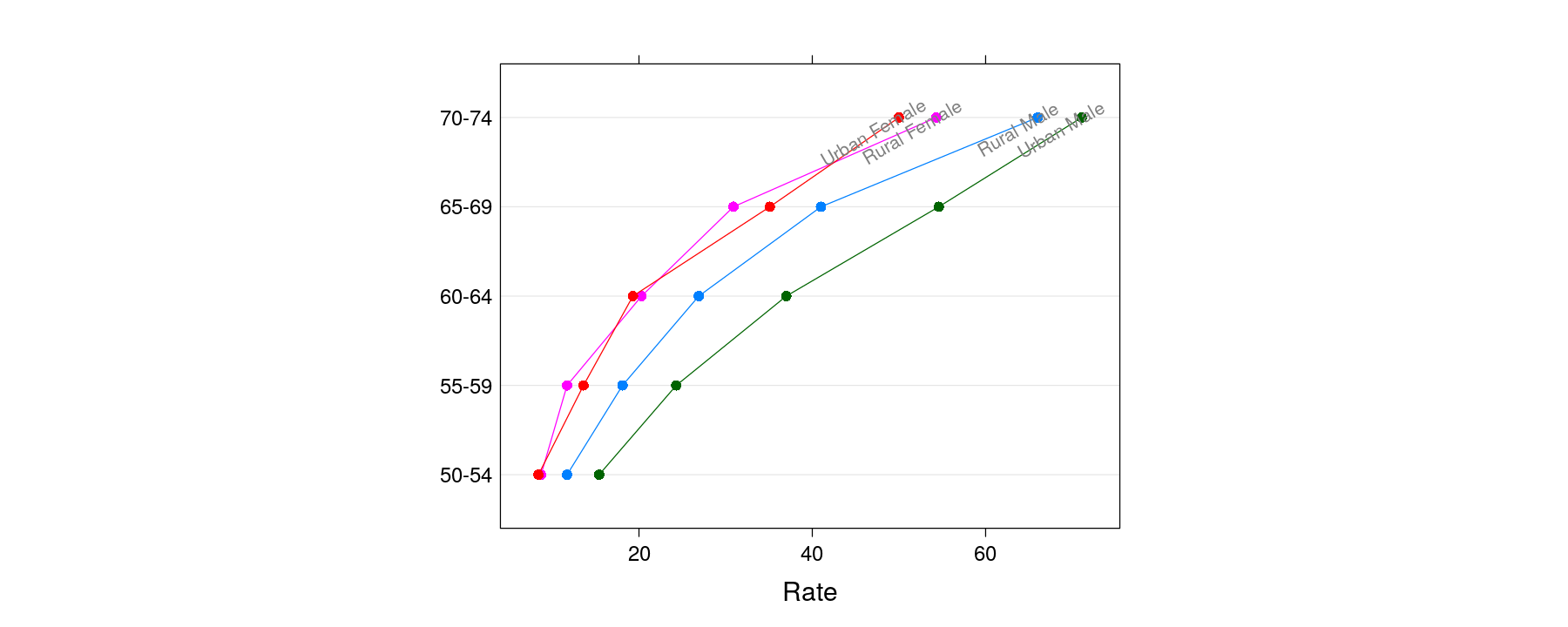
More examples of layering
p <- bwplot(gcsescore ~ gender | factor(score), Chem97, layout = c(6, 1), between = list(x = 0.5))
p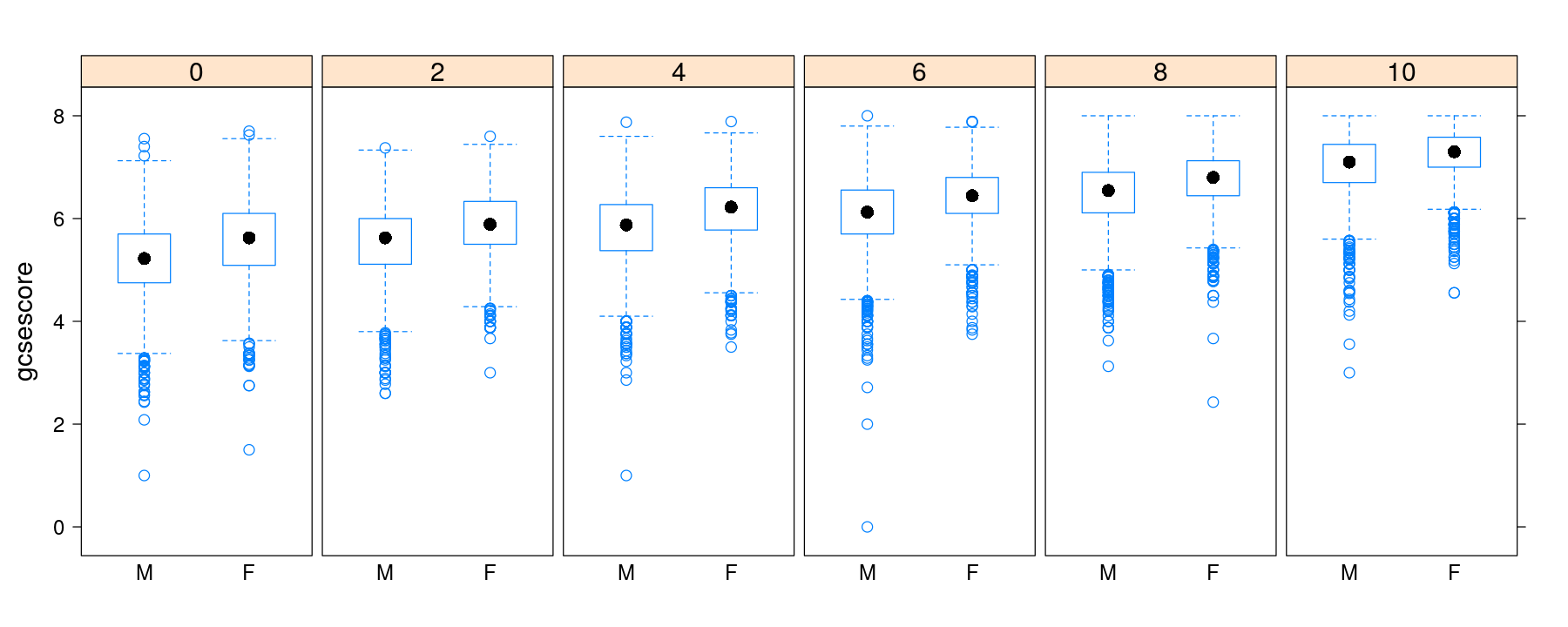
More examples of layering
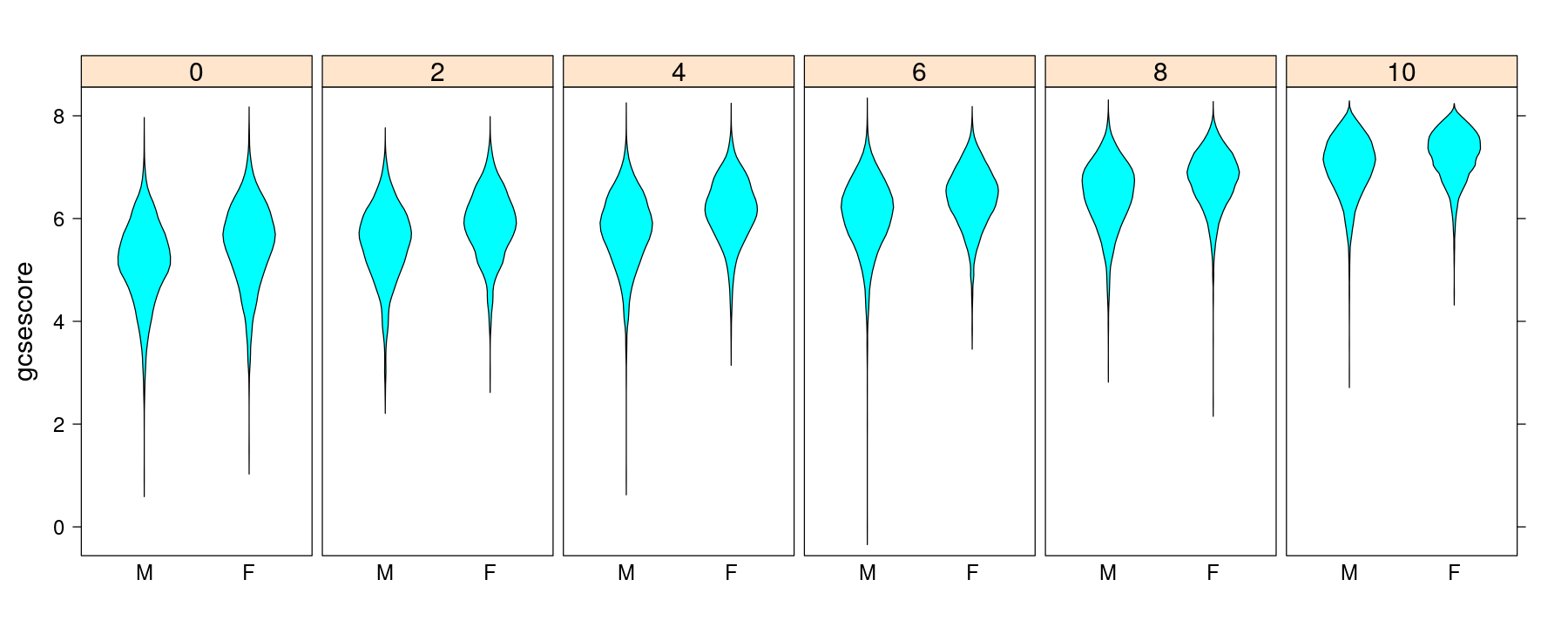
More examples of layering

Custom panel functions
Layers are convenient, but does not completely replace need to write panel functions
Example: smoother with bootstrap confidence intervals
Custom panel functions
data(Anscombe, package = "carData")
xyplot(education ~ income + young + urban, data = Anscombe, outer = TRUE, strip = FALSE,
scales = list(x = list(relation = "free")), between = list(x = 1), layout = c(3, 1),
xlab = c("income", "young", "urban"), panel = panel.smoother.bs, method = "loess")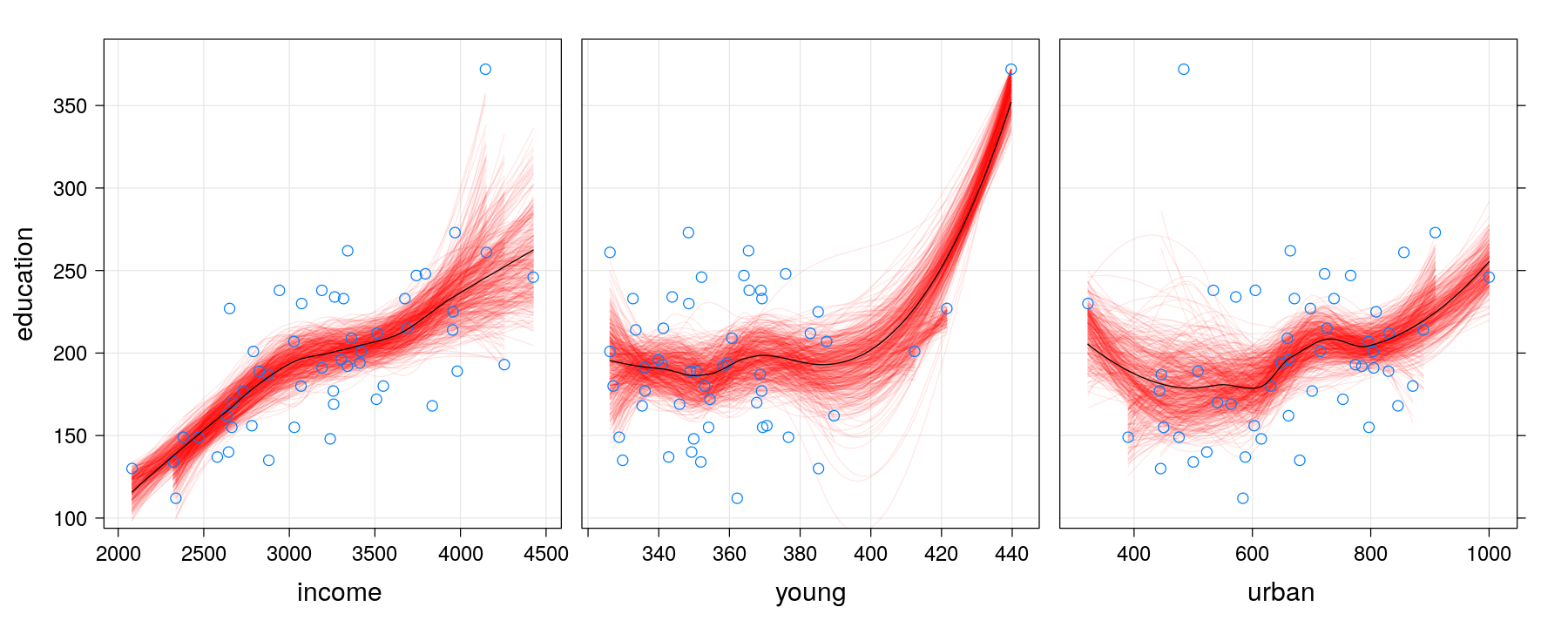
Custom panel functions
xyplot(education ~ income + young + urban, data = Anscombe, outer = TRUE,
scales = list(x = list(relation = "free")), between = list(x = 1), strip = FALSE,
xlab = c("income", "young", "urban"), layout = c(3, 1),
panel = panel.smoother.bs, method = "lm", form = y ~ poly(x, 2), B = 1000)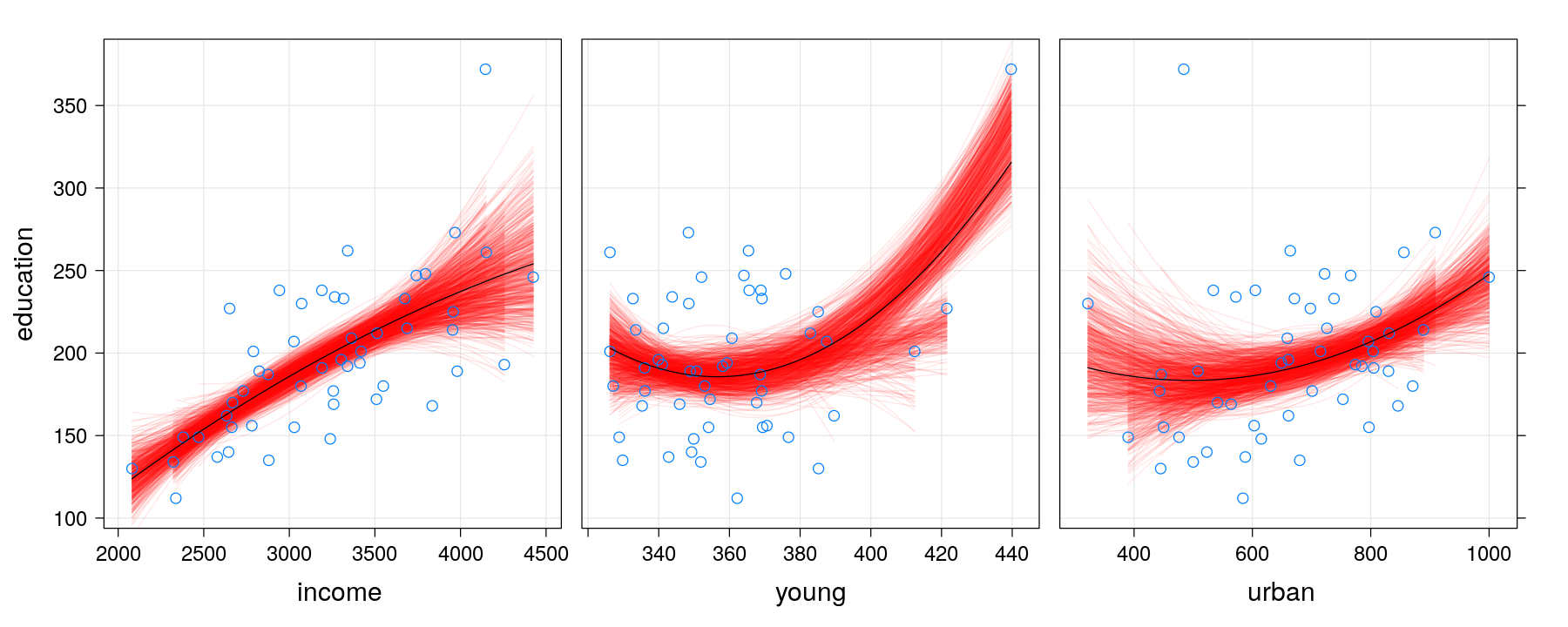
Using subscripts to incorporate additional variables
The
groupsargument allows a categorical variable to define colorSometimes we want to do something else, e.g., bubble charts (size of points encodes a variable)
This is facilitated by the
subscriptsmechanismAn additional variable can be passed to panel function (as seen earlier)
However, this will be the full variable, not panel-specific subsets
If requested, the
subscriptsargument will give panel-specific subset
Using subscripts to incorporate additional variables
- Example: panel function for bubble plot
Using subscripts to incorporate additional variables
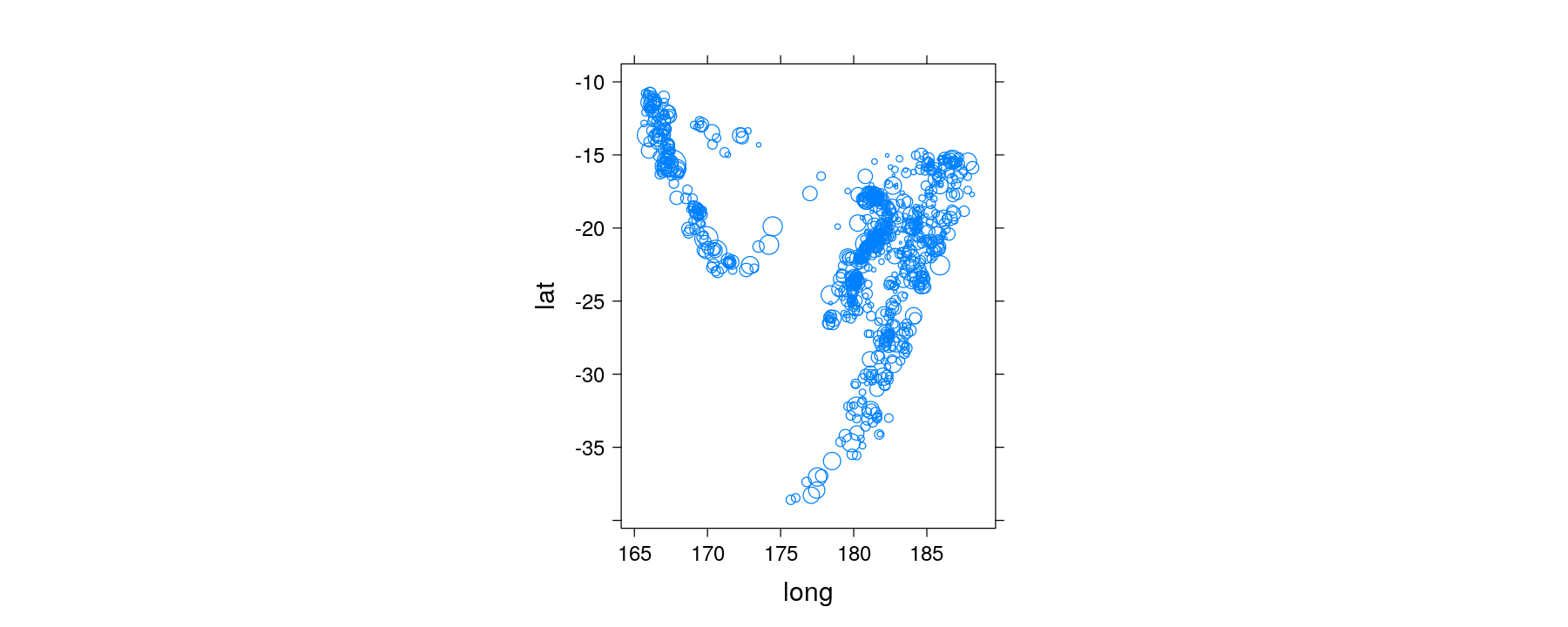
Using subscripts to incorporate additional variables
quakes <- transform(quakes, Depth = equal.count(depth, 6, overlap = 0.1)) # a shingle object
xyplot(lat ~ long | Depth, data = quakes, aspect = "iso",
panel = panel.bubbleplot, size = sqrt(quakes$mag))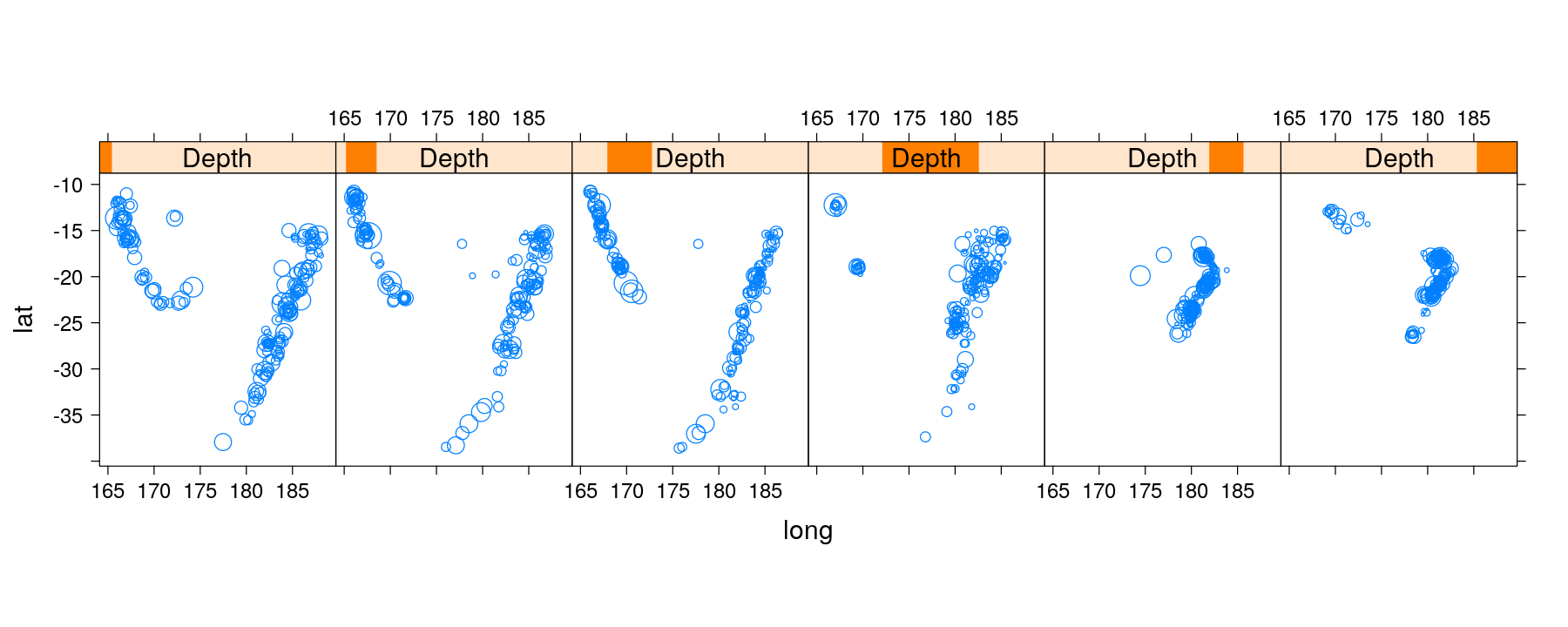
Using subscripts to incorporate additional variables
- A similar function to encode magnitude by color is already implemented
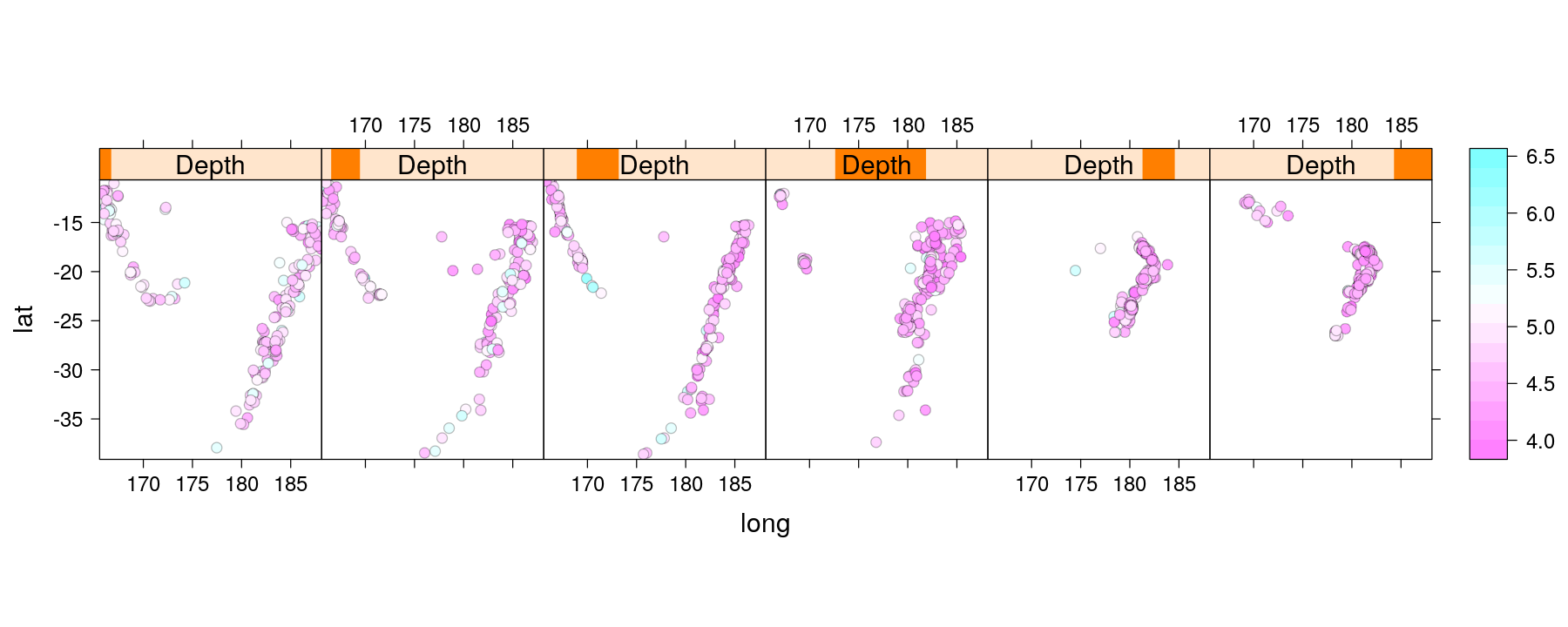
Take-home message
Custom panel functions provide finest level of control
Built-in panel functions are also powerful (but need to read documentation!).
Layering can make life easier (and code easier to read)
Topics we have not discussed in detail
The
prepanel=argument: function that controls axis limits computationsShingle objects: overlapping subintervals defining “levels” for numeric variables
Banking to 45 degrees
Several other high-level functions
S3 methods (dispatch on non-formula objects), e.g.,
barchart,dotplotfor tablesxyplotfor time-series datalevelplot,wireframefor matrices (to display surfaces)
Topics we have not discussed in detail
- Manipulation of “trellis” objects: example
p1 <- barchart(VADeaths, layout = c(NA, 1), groups = FALSE)
p2 <- dotplot(VADeaths, type = "o", par.settings = simpleTheme(pch = 16),
auto.key = list(columns = 2))
print(p1, position = c(0, 0, 0.6, 1))
print(p2, position = c(0.6, 0, 1, 1), newpage = FALSE)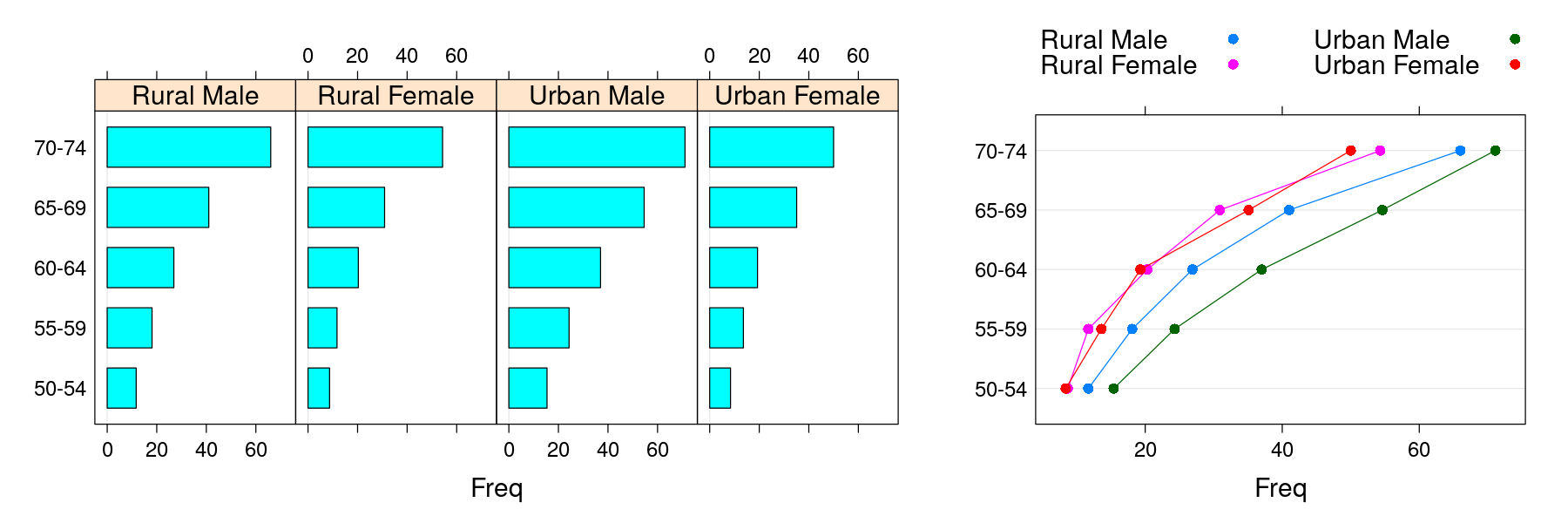
Topics we have not discussed in detail
$Var2
[1] "Rural Male" "Rural Female" "Urban Male" "Urban Female"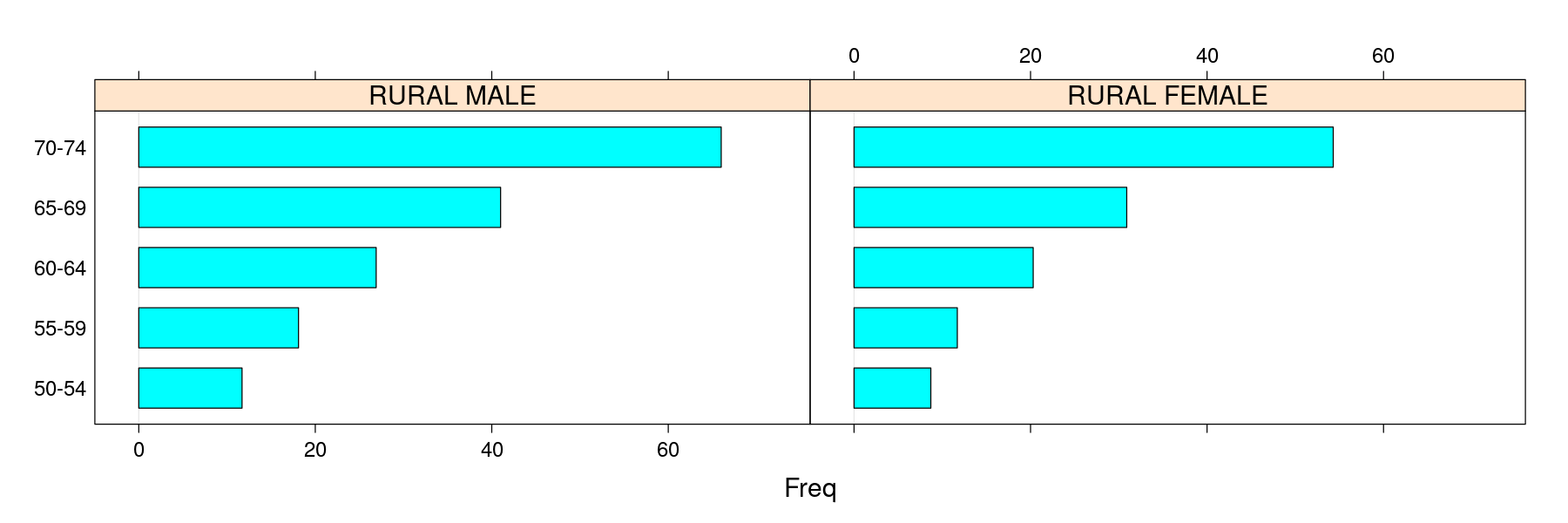
Further reading
The full system would take too long to describe
Online documentation has details; start with
?LatticeLattice book: http://lmdvr.r-forge.r-project.org
Exercises: Regression diagnostics
The goal of the final series of exercises is to recreate some diagnostic plots using lattice
These are modeled on plots and functions in Fox (2015) and the car package
Residual plots
library(package = "car")
Prestige$type <- factor(Prestige$type, levels=c("bc", "wc", "prof"))
fm.prestige <- lm(prestige ~ education + income + type, data = Prestige)
residualPlots(fm.prestige, layout = c(1, 4), tests = FALSE)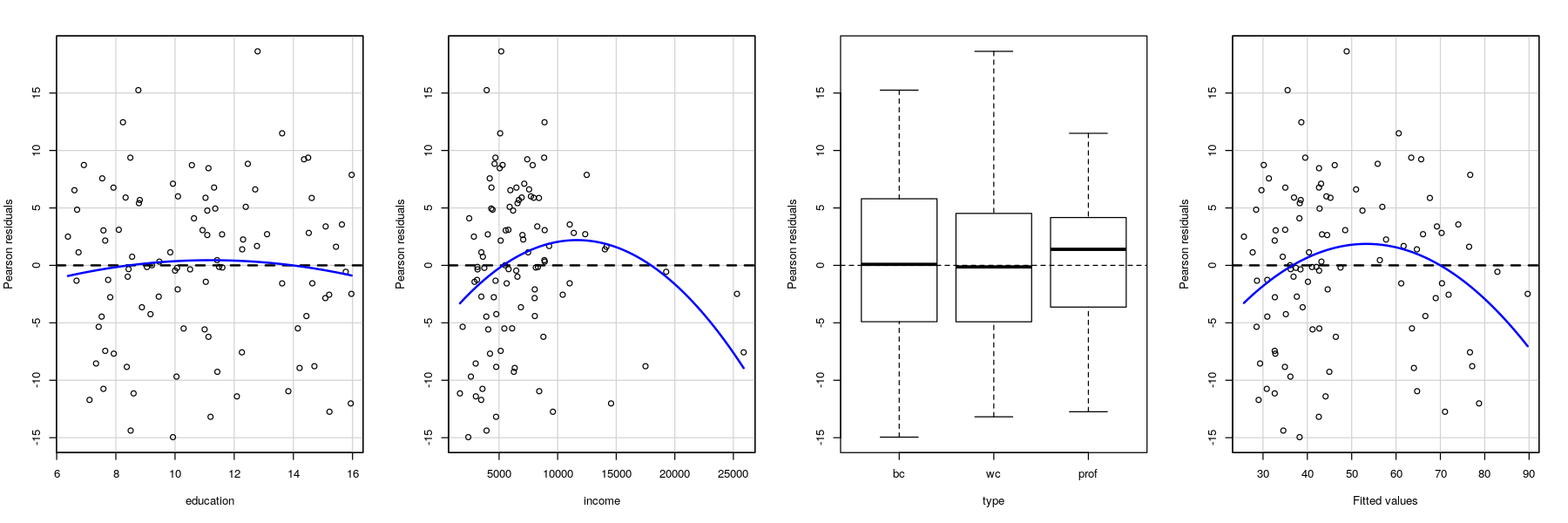
Marginal model plots
## lines are loess smooths of response (Data) and fitted (Model) vs x-variable
marginalModelPlots(fm.prestige, terms = ~ ., fitted = TRUE, layout = c(1, 3), ylab = "prestige")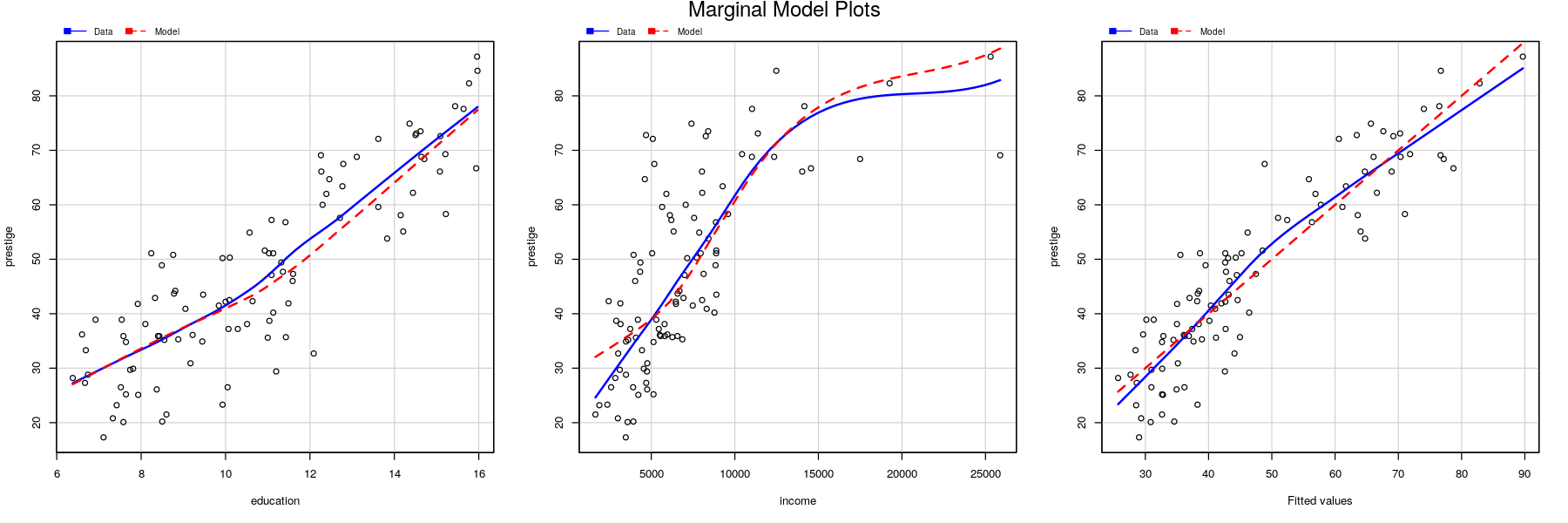
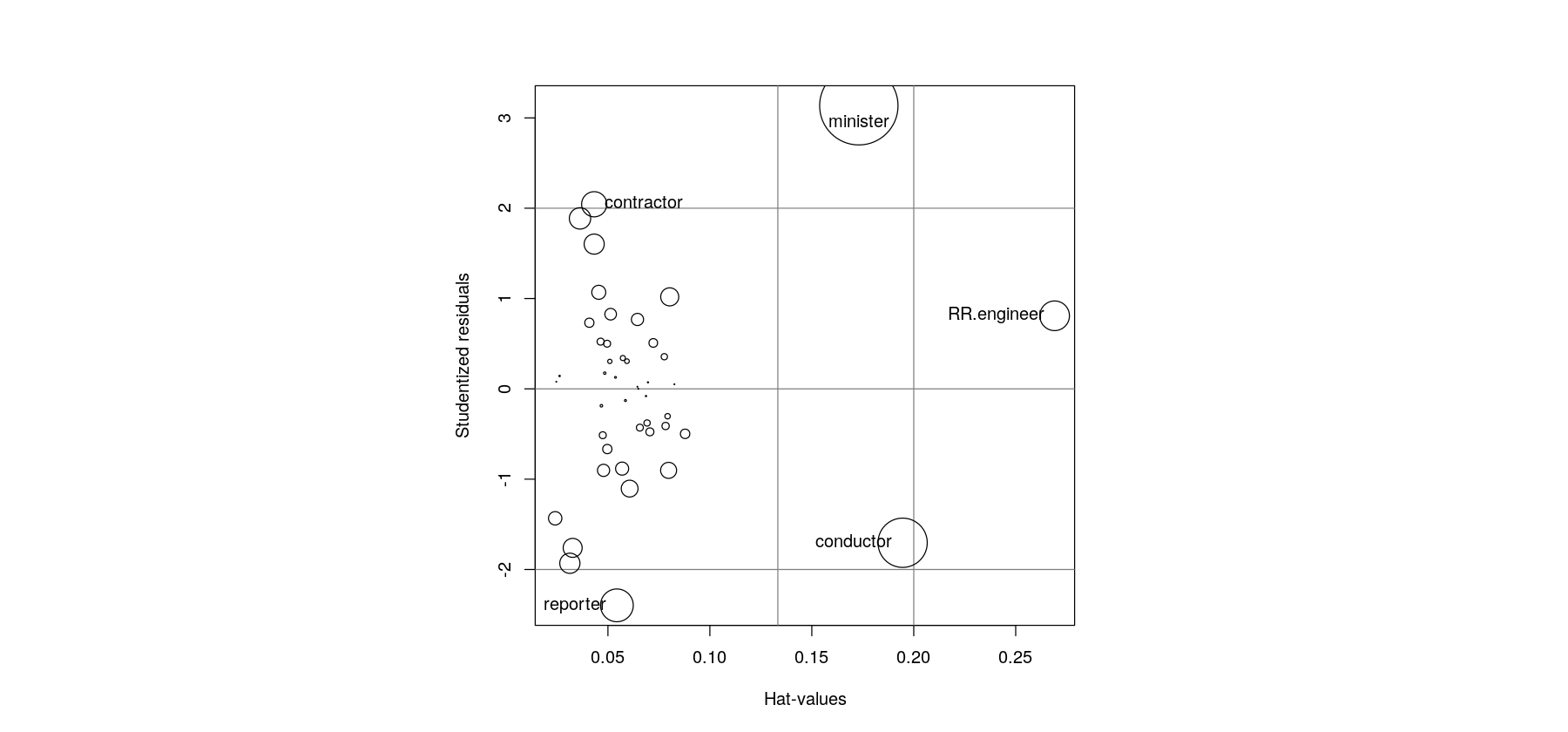
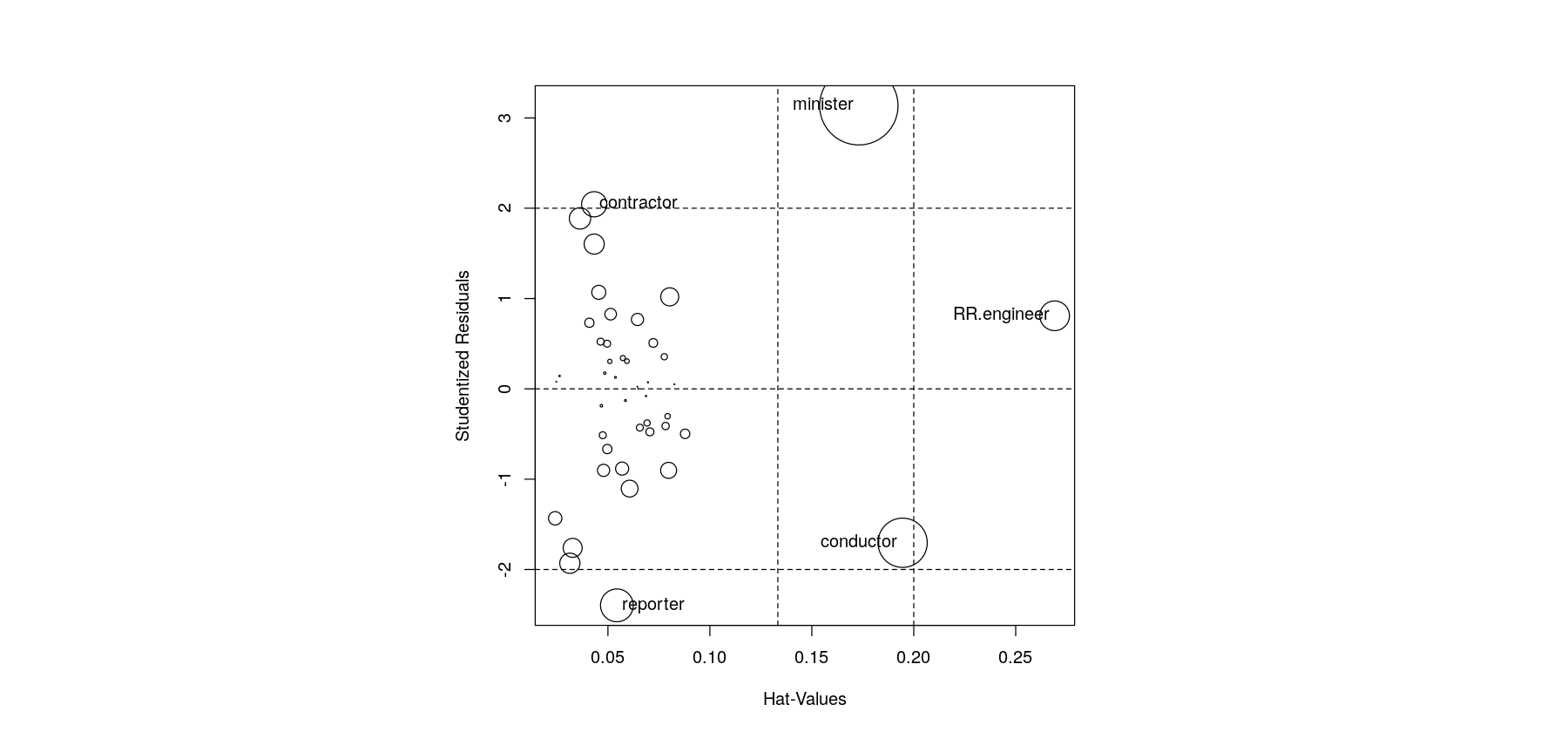
Hat-values, Cook’s distance, Studentized residuals
- Some standard diagnostic measures
fm.duncan <- lm(prestige ~ income + education, data=Duncan)
h <- hatvalues(fm.duncan)
D <- cooks.distance(fm.duncan)
e <- rstudent(fm.duncan)The plot to be recreated is a bubble chart with
- Hat-values on the x-axis
- Studentized residuals on the y-axis
- Points with area proportional to Cook’s distance
- “Unusual” values flagged with text labels
par(pty = "s")
plot(h, e, type = "n", xlab = "Hat-values", ylab = "Studentized residuals", pty = "s")
abline(h = c(-2, 0, 2), v = c(2,3)*mean(h), col = "grey50")
points(h, e, cex = 10 * sqrt(D / max(D)))
id <- abs(e) > 2 | h > 2 * mean(h)
text(h[id], e[id], rownames(Duncan)[id], pos = c(1, 2, 2, 4, 2))
Hat-values, Cook’s distance, Studentized residuals
Hat-values, Cook’s distance, Studentized residuals
Hat-values, Cook’s distance, Studentized residuals
References
Fox, John. 2015. Applied Regression Analysis and Generalized Linear Models. Sage Publications.